Analysis of scRNA-seq & CITE-seq Data Combined
Sub-cluster Macrophages
Jovana Maksimovic
June 21, 2022
Last updated: 2022-06-21
Checks: 7 0
Knit directory:
paed-cf-cite-seq/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20210524) was run prior to running the code in the R Markdown file.
Setting a seed ensures that any results that rely on randomness, e.g.
subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 14ec446. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the
analysis have been committed to Git prior to generating the results (you can
use wflow_publish or wflow_git_commit). workflowr only
checks the R Markdown file, but you know if there are other scripts or data
files that it depends on. Below is the status of the Git repository when the
results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/obsolete/
Ignored: code/obsolete/
Ignored: data/190930_A00152_0150_BHTYCMDSXX/
Ignored: data/CellRanger/
Ignored: data/GSE127465_RAW/
Ignored: data/SCEs/02_ZILIONIS.sct_normalised.SEU.rds
Ignored: data/SCEs/03_C133_Neeland.demultiplexed.SCE.rds
Ignored: data/SCEs/03_C133_Neeland.emptyDrops.SCE.rds
Ignored: data/SCEs/03_C133_Neeland.preprocessed.SCE.rds
Ignored: data/SCEs/03_CF_BAL_Pilot.CellRanger_v6.SCE.rds
Ignored: data/SCEs/03_CF_BAL_Pilot.emptyDrops.SCE.rds
Ignored: data/SCEs/03_CF_BAL_Pilot.preprocessed.SCE.rds
Ignored: data/SCEs/03_COMBO.clustered.SEU.rds
Ignored: data/SCEs/03_COMBO.clustered_annotated_macrophages_diet.SEU.rds
Ignored: data/SCEs/03_COMBO.clustered_annotated_others_diet.SEU.rds
Ignored: data/SCEs/03_COMBO.clustered_annotated_tcells_diet.SEU.rds
Ignored: data/SCEs/03_COMBO.clustered_diet.SEU.rds
Ignored: data/SCEs/03_COMBO.integrated.SEU.rds
Ignored: data/SCEs/03_COMBO.zilionis_mapped.SEU.rds
Ignored: data/SCEs/04_C133_Neeland.adt_dsb_normalised.rds
Ignored: data/SCEs/04_C133_Neeland.adt_integrated.rds
Ignored: data/SCEs/04_C133_Neeland.all_integrated.SEU.rds
Ignored: data/SCEs/04_CF_BAL_Pilot.CellRanger_v6.SCE.rds
Ignored: data/SCEs/04_CF_BAL_Pilot.emptyDrops.SCE.rds
Ignored: data/SCEs/04_CF_BAL_Pilot.preprocessed.SCE.rds
Ignored: data/SCEs/04_CF_BAL_Pilot.transfer_adt.SEU.rds
Ignored: data/SCEs/04_COMBO.clean_clustered.SEU.rds
Ignored: data/SCEs/04_COMBO.clean_clustered.SEU_bk.rds
Ignored: data/SCEs/04_COMBO.clean_integrated.SEU.rds
Ignored: data/SCEs/04_COMBO.clean_integrated.SEU_bk.rds
Ignored: data/SCEs/04_COMBO.clean_macrophages_diet.SEU.rds
Ignored: data/SCEs/04_COMBO.clean_others_diet.SEU.rds
Ignored: data/SCEs/04_COMBO.clean_tcells_diet.SEU.rds
Ignored: data/SCEs/04_COMBO.clustered.SEU.rds
Ignored: data/SCEs/04_COMBO.clustered_annotated_adt_diet.SEU.rds
Ignored: data/SCEs/04_COMBO.clustered_annotated_lung_diet.SEU.rds
Ignored: data/SCEs/04_COMBO.clustered_annotated_macrophages_diet.SEU.rds
Ignored: data/SCEs/04_COMBO.clustered_annotated_others_diet.SEU.rds
Ignored: data/SCEs/04_COMBO.clustered_annotated_tcells_diet.SEU.rds
Ignored: data/SCEs/04_COMBO.clustered_diet.SEU.rds
Ignored: data/SCEs/04_COMBO.integrated.SEU.rds
Ignored: data/SCEs/04_COMBO.macrophages_clustered.SEU.rds
Ignored: data/SCEs/04_COMBO.macrophages_integrated.SEU.rds
Ignored: data/SCEs/04_COMBO.others_clustered.SEU.rds
Ignored: data/SCEs/04_COMBO.others_integrated.SEU.rds
Ignored: data/SCEs/04_COMBO.tcells_clustered.SEU.rds
Ignored: data/SCEs/04_COMBO.tcells_integrated.SEU.rds
Ignored: data/SCEs/04_COMBO.zilionis_mapped.SEU.rds
Ignored: data/SCEs/05_CF_BAL_Pilot.transfer_adt.SEU.rds
Ignored: data/SCEs/05_COMBO.clean_clustered.SEU.rds
Ignored: data/SCEs/05_COMBO.clean_integrated.SEU.rds
Ignored: data/SCEs/05_COMBO.clean_macrophages_diet.SEU.rds
Ignored: data/SCEs/05_COMBO.clean_others_diet.SEU.rds
Ignored: data/SCEs/05_COMBO.clean_tcells_diet.SEU.rds
Ignored: data/SCEs/05_COMBO.clustered_annotated_adt_diet.SEU.rds
Ignored: data/SCEs/05_COMBO.clustered_annotated_lung_diet.SEU.rds
Ignored: data/SCEs/05_COMBO.clustered_annotated_macrophages_diet.SEU.rds
Ignored: data/SCEs/05_COMBO.clustered_annotated_others_diet.SEU.rds
Ignored: data/SCEs/05_COMBO.clustered_annotated_tcells_diet.SEU.rds
Ignored: data/SCEs/05_COMBO.macrophages_clustered.SEU.rds
Ignored: data/SCEs/05_COMBO.macrophages_integrated.SEU.rds
Ignored: data/SCEs/05_COMBO.others_clustered.SEU.rds
Ignored: data/SCEs/05_COMBO.others_integrated.SEU.rds
Ignored: data/SCEs/05_COMBO.tcells_clustered.SEU.rds
Ignored: data/SCEs/05_COMBO.tcells_integrated.SEU.rds
Ignored: data/SCEs/06_COMBO.clean_clustered.SEU.rds
Ignored: data/SCEs/06_COMBO.clean_integrated.SEU.rds
Ignored: data/SCEs/06_COMBO.clean_macrophages_diet.SEU.rds
Ignored: data/SCEs/06_COMBO.clean_others_diet.SEU.rds
Ignored: data/SCEs/06_COMBO.clean_tcells_diet.SEU.rds
Ignored: data/SCEs/06_COMBO.macrophages_clustered.SEU.rds
Ignored: data/SCEs/06_COMBO.macrophages_integrated.SEU.rds
Ignored: data/SCEs/06_COMBO.others_clustered.SEU.rds
Ignored: data/SCEs/06_COMBO.others_integrated.SEU.rds
Ignored: data/SCEs/06_COMBO.tcells_clustered.SEU.rds
Ignored: data/SCEs/06_COMBO.tcells_integrated.SEU.rds
Ignored: data/SCEs/C133_Neeland.CellRanger.SCE.rds
Ignored: data/SCEs/obsolete/
Ignored: data/cellsnp-lite/
Ignored: data/emptyDrops/obsolete/
Ignored: data/obsolete/
Ignored: data/sample_sheets/obsolete/
Ignored: output/marker-analysis/obsolete/
Ignored: output/obsolete/
Ignored: rename_captures.R
Ignored: renv/library/
Ignored: renv/staging/
Ignored: wflow_background.R
Unstaged changes:
Modified: .gitignore
Modified: .renvignore
Modified: analysis/ref.bib
Modified: renv/.gitignore
Modified: renv/settings.dcf
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made
to the R Markdown (analysis/08_COMBO.cluster_macrophages.Rmd) and HTML (docs/08_COMBO.cluster_macrophages.html)
files. If you’ve configured a remote Git repository (see
?wflow_git_remote), click on the hyperlinks in the table below to
view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 14ec446 | Jovana Maksimovic | 2022-06-21 | wflow_publish(c("analysis/08_COMBO.cluster_macrophages.Rmd", |
| Rmd | f3b7b92 | Jovana Maksimovic | 2022-06-16 | Submission version |
| html | f3b7b92 | Jovana Maksimovic | 2022-06-16 | Submission version |
1 Load libraries
2 Load Data
Load the clustered and labelled scRNA-seq and CITE-seq data.
seu <- readRDS(file = here("data/SCEs/05_COMBO.clustered_annotated_macrophages_diet.SEU.rds"))
DefaultAssay(seu) <- "RNA"
entrez <- select(org.Hs.eg.db, columns = c("ENTREZID","SYMBOL"),
keys = keys(org.Hs.eg.db))
entrez <- entrez[!is.na(entrez$ENTREZID),]
seu <- seu[rownames(seu) %in% entrez$SYMBOL,]3 Subcluster macrophages
Normalise and integrate data.
out <- here("data/SCEs/06_COMBO.macrophages_integrated.SEU.rds")
if(!file.exists(out)){
seuInt <- intDat(seu, type = "RNA",
reference = unique(seu$capture[seu$experiment == 1]))
saveRDS(seuInt, file = out)
} else {
seuInt <- readRDS(file = out)
}Visualise the data.
seuInt <- RunPCA(seuInt, verbose = FALSE, dims = 1:30) %>%
RunUMAP(verbose = FALSE, dims = 1:30)DimPlot(seuInt, group.by = "experiment", combine = FALSE)[[1]]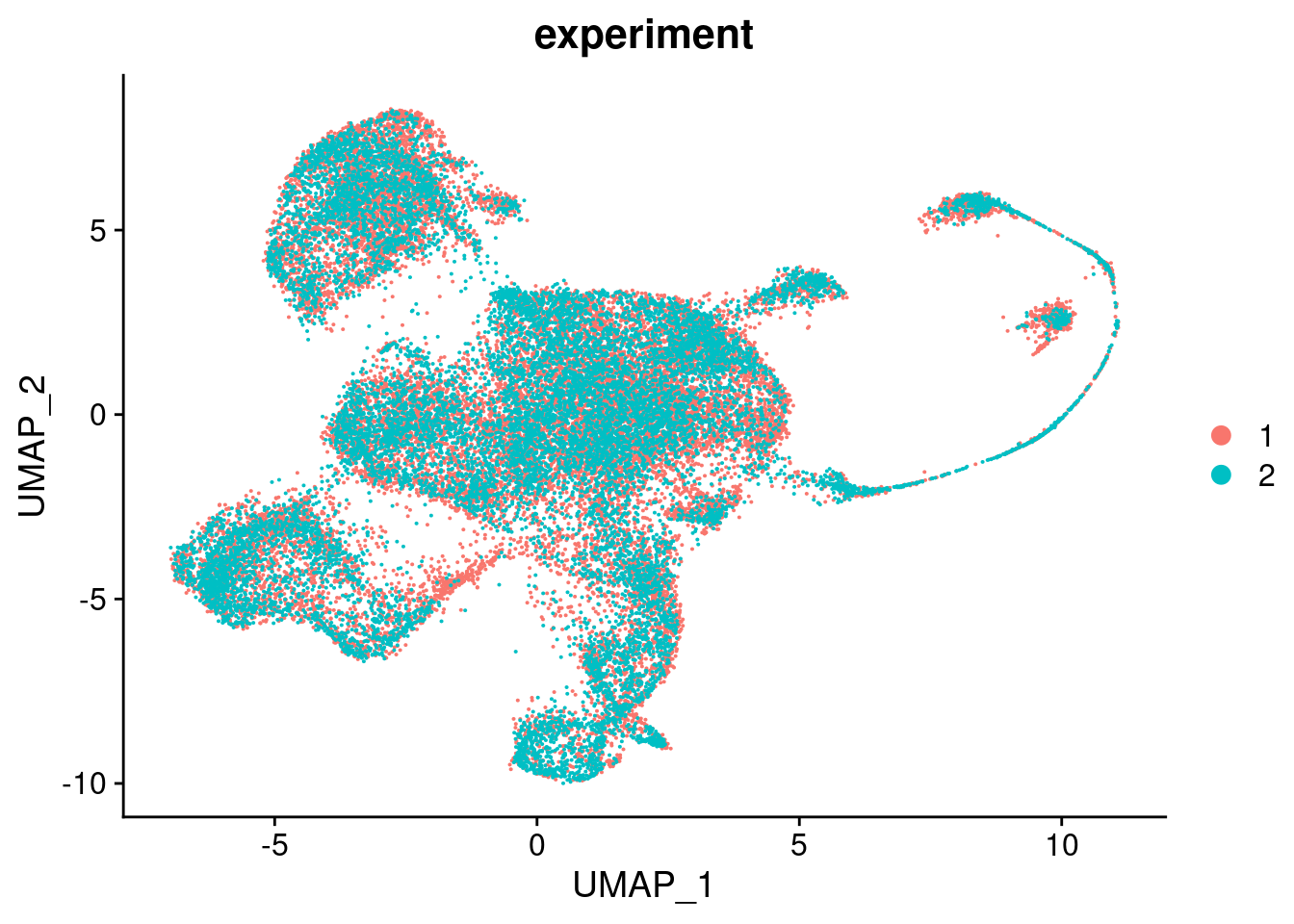
| Version | Author | Date |
|---|---|---|
| f3b7b92 | Jovana Maksimovic | 2022-06-16 |
4 Clustering
4.1 Perform Linear Dimensional Reduction
p1 <- DimPlot(seuInt, reduction = "pca", group.by = "donor")
p2 <- DimPlot(seuInt, reduction = "pca", dims = c(1,3), group.by = "donor")
p3 <- DimPlot(seuInt, reduction = "pca", dims = c(2,3), group.by = "donor")
p4 <- DimPlot(seuInt, reduction = "pca", dims = c(3,4), group.by = "donor")
((p1 | p2) / (p3 | p4)) + plot_layout(guides = "collect") &
theme(legend.text = element_text(size = 8),
plot.title = element_text(size = 10),
axis.title = element_text(size = 9),
axis.text = element_text(size = 8))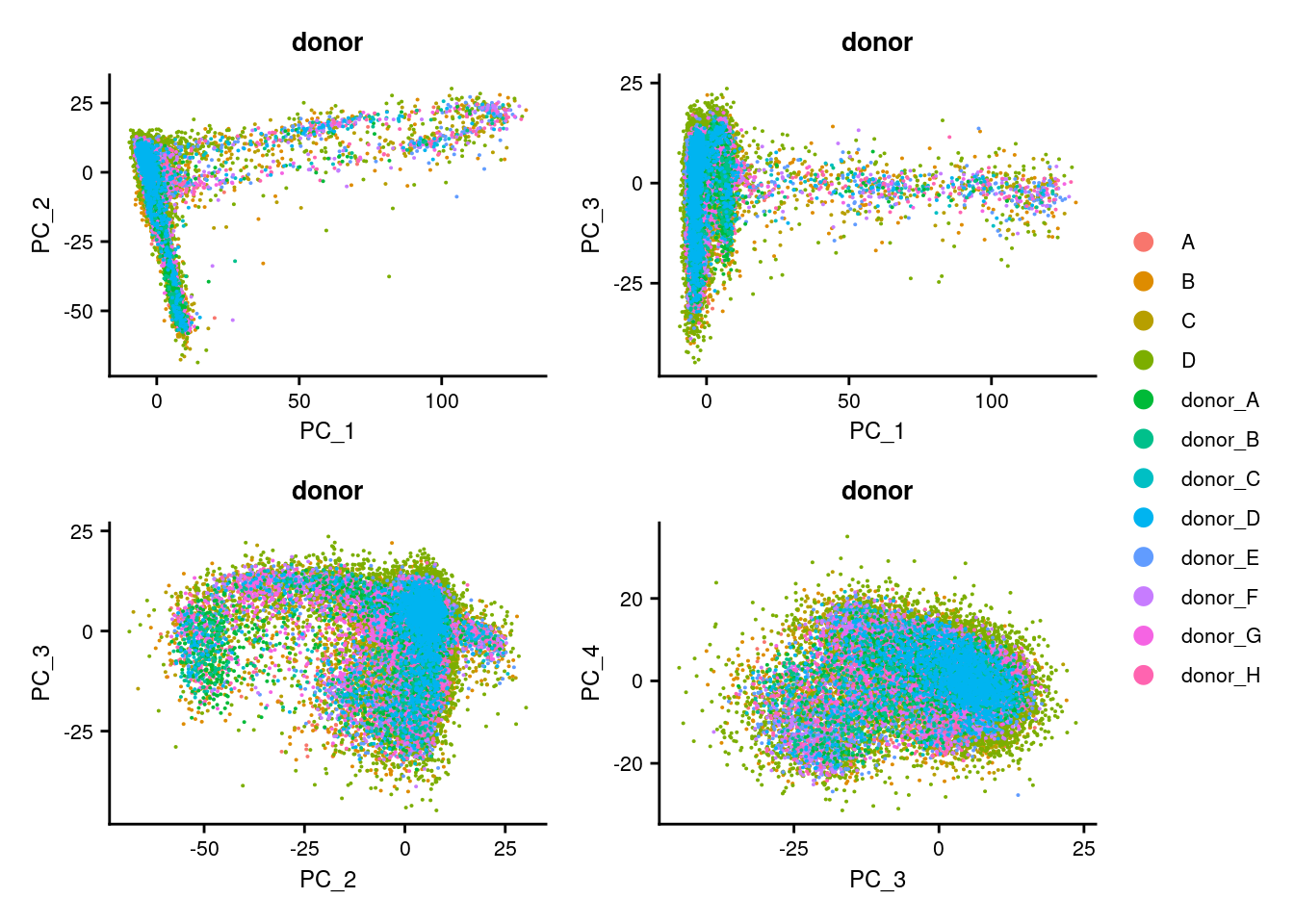
| Version | Author | Date |
|---|---|---|
| f3b7b92 | Jovana Maksimovic | 2022-06-16 |
DimHeatmap(seuInt, dims = 1:30, cells = 500, balanced = TRUE)
| Version | Author | Date |
|---|---|---|
| f3b7b92 | Jovana Maksimovic | 2022-06-16 |
4.2 Determine the ‘Dimensionality’ of the Dataset
ElbowPlot(seuInt, ndims = 30)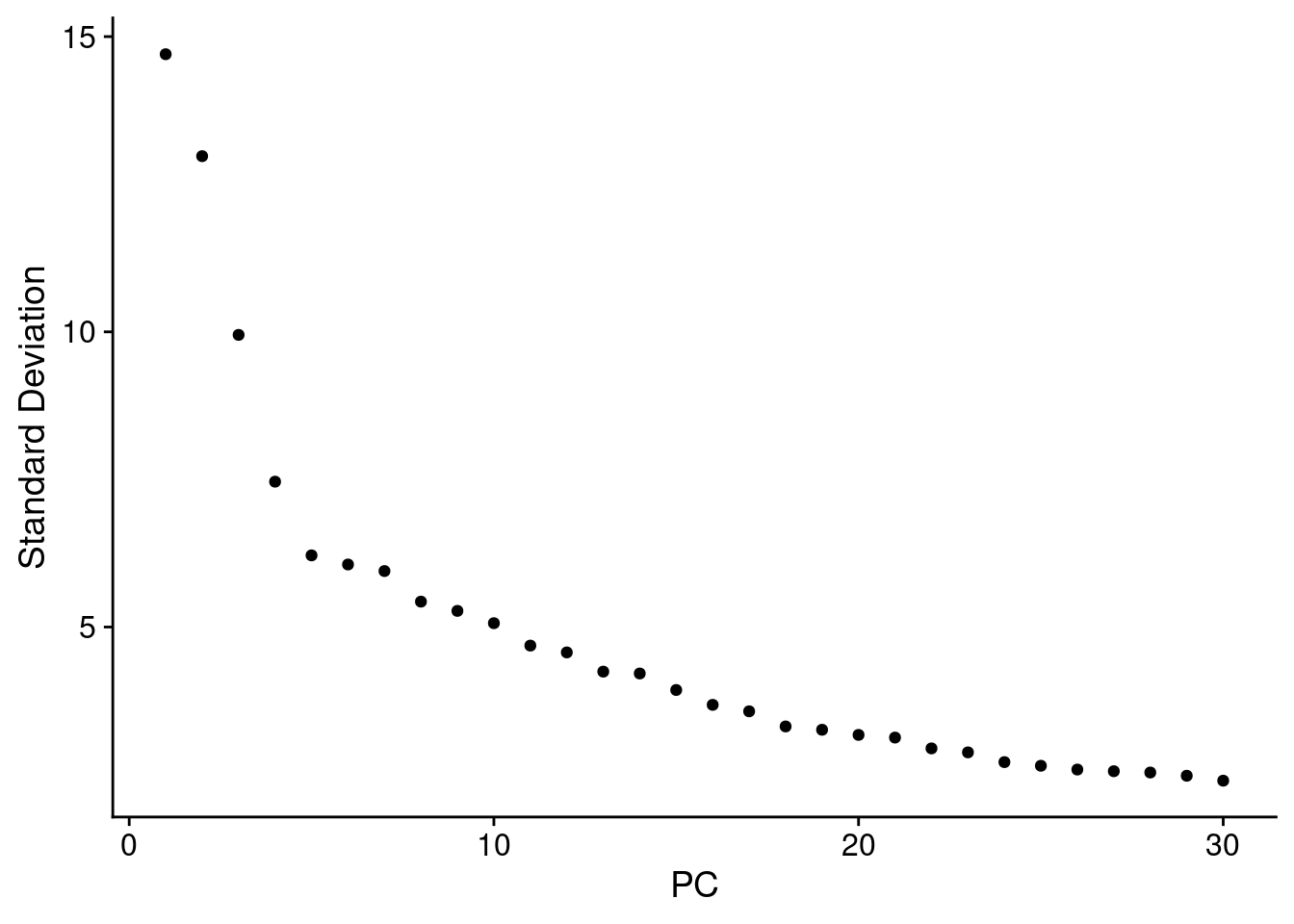
| Version | Author | Date |
|---|---|---|
| f3b7b92 | Jovana Maksimovic | 2022-06-16 |
5 Cluster the Cells
Examine cluster number and size with respect to resolution.
out <- here("data/SCEs/06_COMBO.macrophages_clustered.SEU.rds")
if(!file.exists(out)){
seuInt <- FindNeighbors(seuInt, reduction = "pca", dims = 1:30)
seuInt <- FindClusters(seuInt, algorithm = 3,
resolution = seq(0.1, 1, by = 0.1))
seuInt <- RunUMAP(seuInt, dims = 1:10)
saveRDS(seuInt, file = out)
} else {
seuInt <- readRDS(file = out)
}
clustree::clustree(seuInt)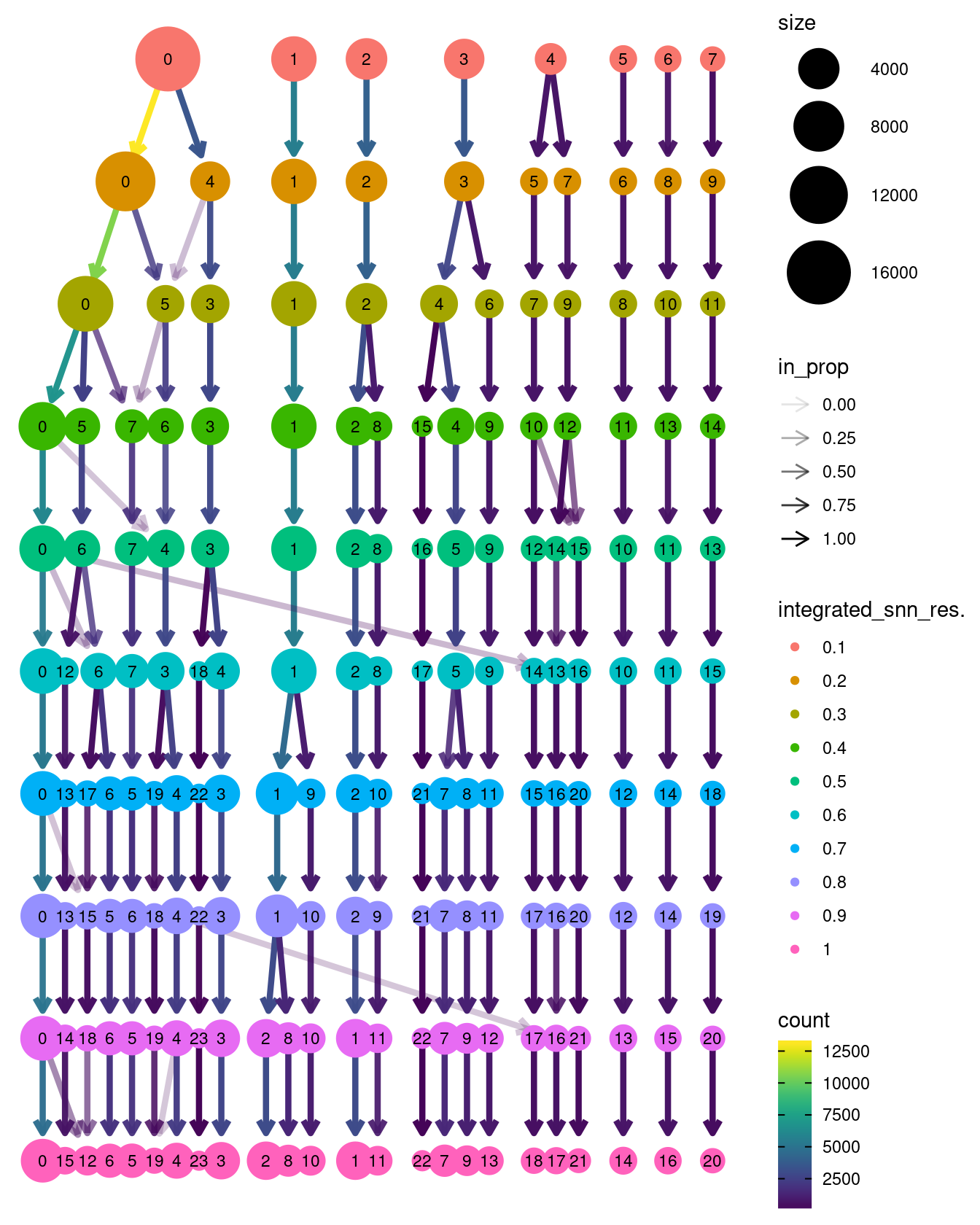
| Version | Author | Date |
|---|---|---|
| f3b7b92 | Jovana Maksimovic | 2022-06-16 |
Choose a resolution. Visualise UMAP.
options(ggrepel.max.overlaps = Inf)
grp <- "integrated_snn_res.1"
DimPlot(seuInt, reduction = 'umap', label = TRUE, repel = FALSE,
label.size = 2.5, group.by = grp) + NoLegend()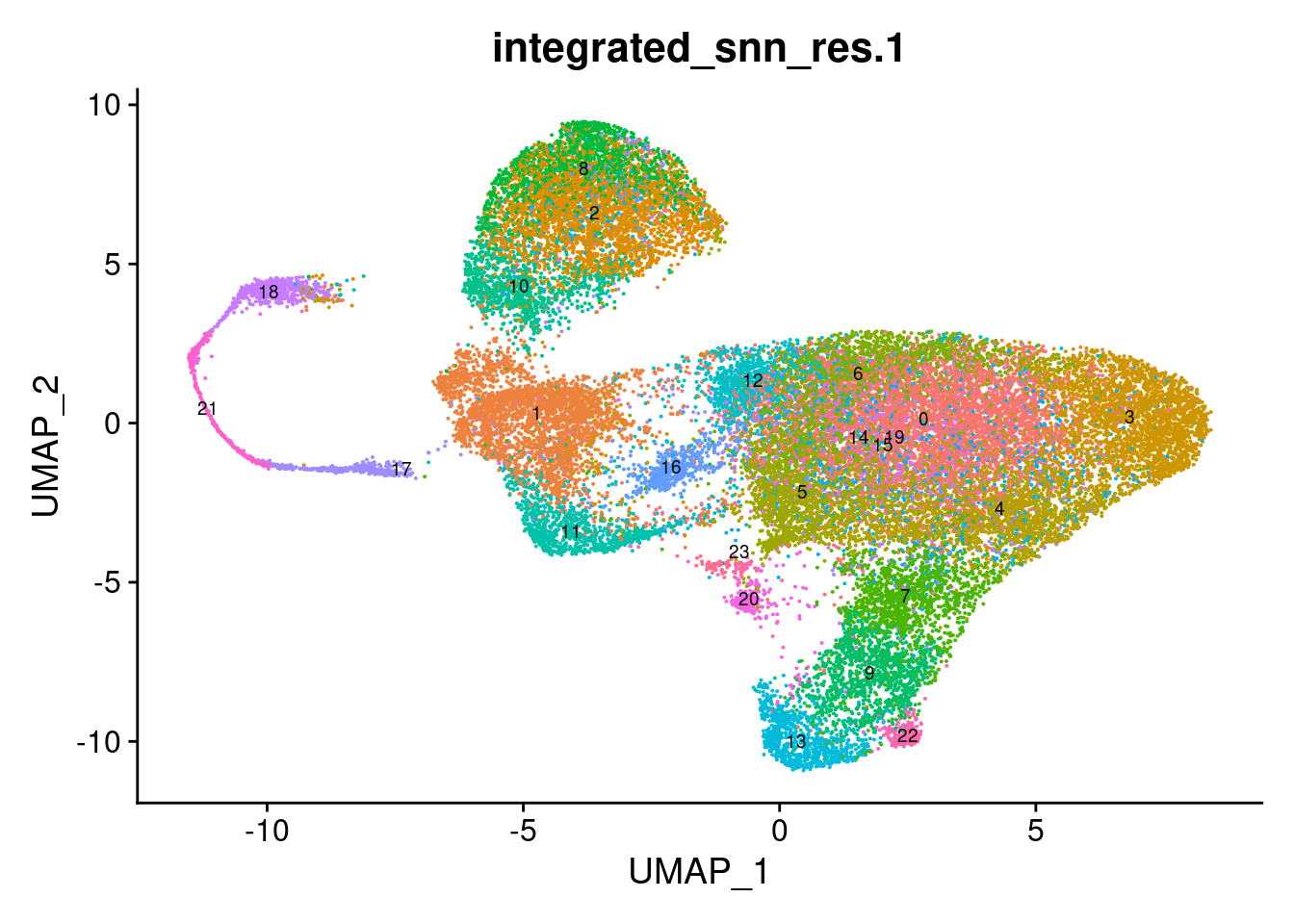
| Version | Author | Date |
|---|---|---|
| f3b7b92 | Jovana Maksimovic | 2022-06-16 |
5.1 Examine clusters
Visualise quality metrics by cluster.
seuInt@meta.data %>%
ggplot(aes(x = integrated_snn_res.1,
y = predicted.annotation.l1.score,
fill = integrated_snn_res.1)) +
geom_violin(scale = "width") +
NoLegend() -> p1
seuInt@meta.data %>%
ggplot(aes(x = integrated_snn_res.1,
y = nCount_RNA,
fill = integrated_snn_res.1)) +
geom_violin(scale = "area") +
scale_y_log10() +
NoLegend() -> p2
seuInt@meta.data %>%
ggplot(aes(x = integrated_snn_res.1,
y = nFeature_RNA,
fill = integrated_snn_res.1)) +
geom_violin(scale = "area") +
scale_y_log10() +
NoLegend() -> p3
seuInt@meta.data %>%
ggplot(aes(x = integrated_snn_res.1,
y = predicted.ann_level_3.score,
fill = integrated_snn_res.1)) +
geom_violin(scale = "area") +
scale_y_log10() +
NoLegend() -> p4
((p1 | p2) / (p3 | p4)) & theme(text = element_text(size = 8))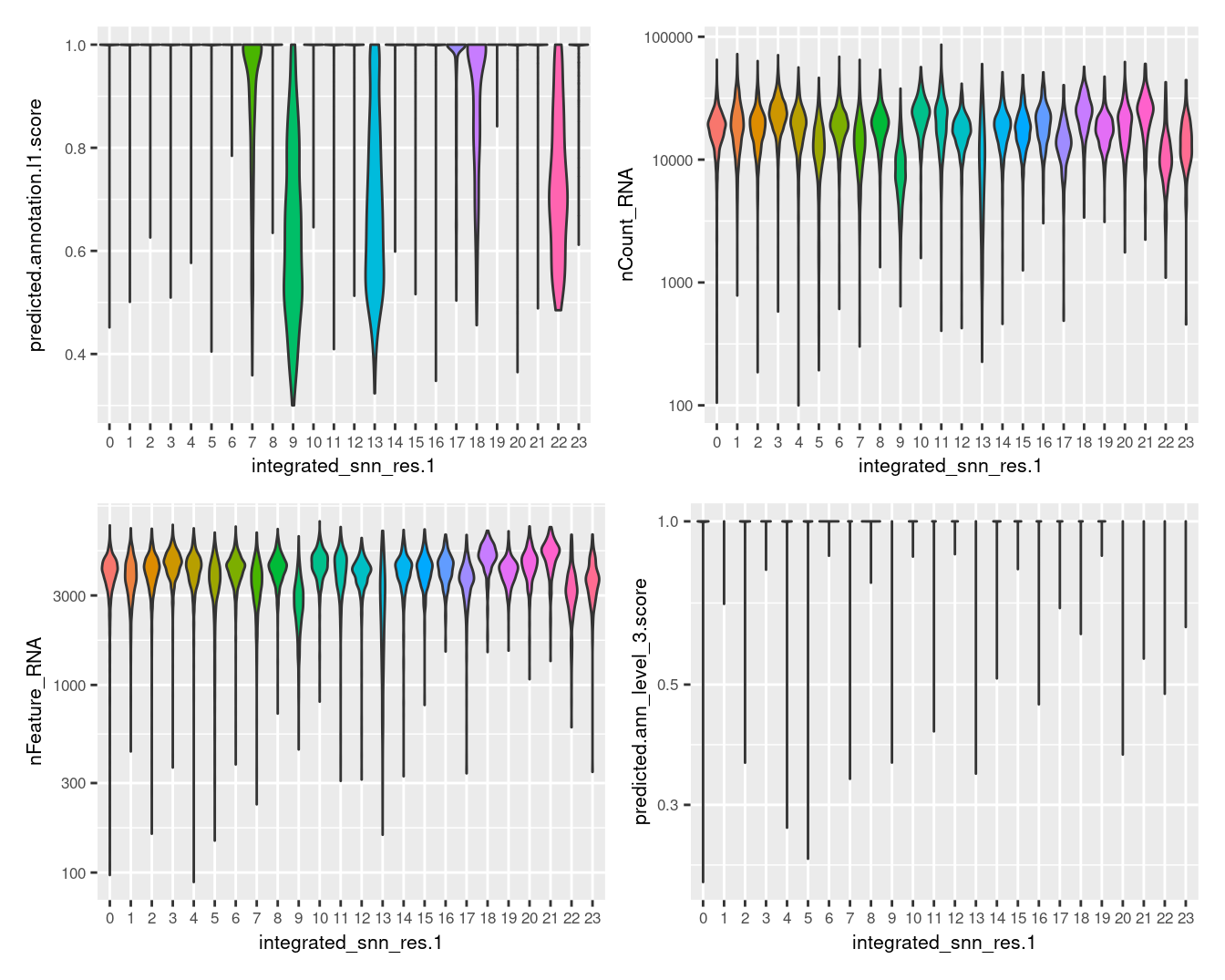
| Version | Author | Date |
|---|---|---|
| f3b7b92 | Jovana Maksimovic | 2022-06-16 |
6 Identify Cluster Marker Genes
Adapted from Dr. Belinda Phipson’s work for (Sim et al. 2021).
6.1 Test for Marker Genes using limma
# limma-trend for DE
Idents(seuInt) <- grp
counts <- as.matrix(seuInt[["RNA"]]@counts)
y.org <- DGEList(counts)
logcounts <- normCounts(y.org, log = TRUE, prior.count = 0.5)
maxclust <- length(levels(Idents(seuInt))) - 1
clustgrp <- paste0("c", Idents(seuInt))
clustgrp <- factor(clustgrp, levels = paste0("c", 0:maxclust))
donor <- seuInt$donor
design <- model.matrix(~ 0 + clustgrp + donor)
colnames(design)[1:(length(levels(clustgrp)))] <- levels(clustgrp)
# Create contrast matrix
mycont <- matrix(NA, ncol = length(levels(clustgrp)),
nrow = length(levels(clustgrp)))
rownames(mycont) <- colnames(mycont) <- levels(clustgrp)
diag(mycont) <- 1
mycont[upper.tri(mycont)] <- -1/(length(levels(factor(clustgrp))) - 1)
mycont[lower.tri(mycont)] <- -1/(length(levels(factor(clustgrp))) - 1)
# Fill out remaining rows with 0s
zero.rows <- matrix(0, ncol = length(levels(clustgrp)),
nrow = (ncol(design) - length(levels(clustgrp))))
fullcont <- rbind(mycont, zero.rows)
rownames(fullcont) <- colnames(design)
fit <- lmFit(logcounts, design)
fit.cont <- contrasts.fit(fit, contrasts = fullcont)
fit.cont <- eBayes(fit.cont, trend = TRUE, robust = TRUE)
summary(decideTests(fit.cont)) c0 c1 c2 c3 c4 c5 c6 c7 c8 c9 c10 c11
Down 5294 7241 2583 4024 2208 4370 2432 4077 2027 7064 2484 3694
NotSig 8279 6781 10589 8449 10752 7078 9895 9953 10611 6484 11200 10721
Up 2005 1556 2406 3105 2618 4130 3251 1548 2940 2030 1894 1163
c12 c13 c14 c15 c16 c17 c18 c19 c20 c21 c22 c23
Down 2904 6800 1689 1417 1304 2416 889 1268 584 1921 2813 1627
NotSig 11321 6506 12822 11736 12614 11150 9993 12749 11762 8507 10583 12534
Up 1353 2272 1067 2425 1660 2012 4696 1561 3232 5150 2182 14176.2 Test relative to a threshold (TREAT)
tr <- treat(fit.cont, fc = 1.5)
dt <- decideTests(tr)
summary(dt) c0 c1 c2 c3 c4 c5 c6 c7 c8 c9 c10 c11
Down 4 54 1 13 2 20 9 42 5 294 5 2
NotSig 15565 15485 15569 15522 15563 15532 15535 15515 15550 15175 15545 15509
Up 9 39 8 43 13 26 34 21 23 109 28 67
c12 c13 c14 c15 c16 c17 c18 c19 c20 c21 c22 c23
Down 6 413 0 2 0 11 1 1 0 60 221 62
NotSig 15550 15012 15569 15547 15503 15534 15355 15547 15530 14903 15134 15495
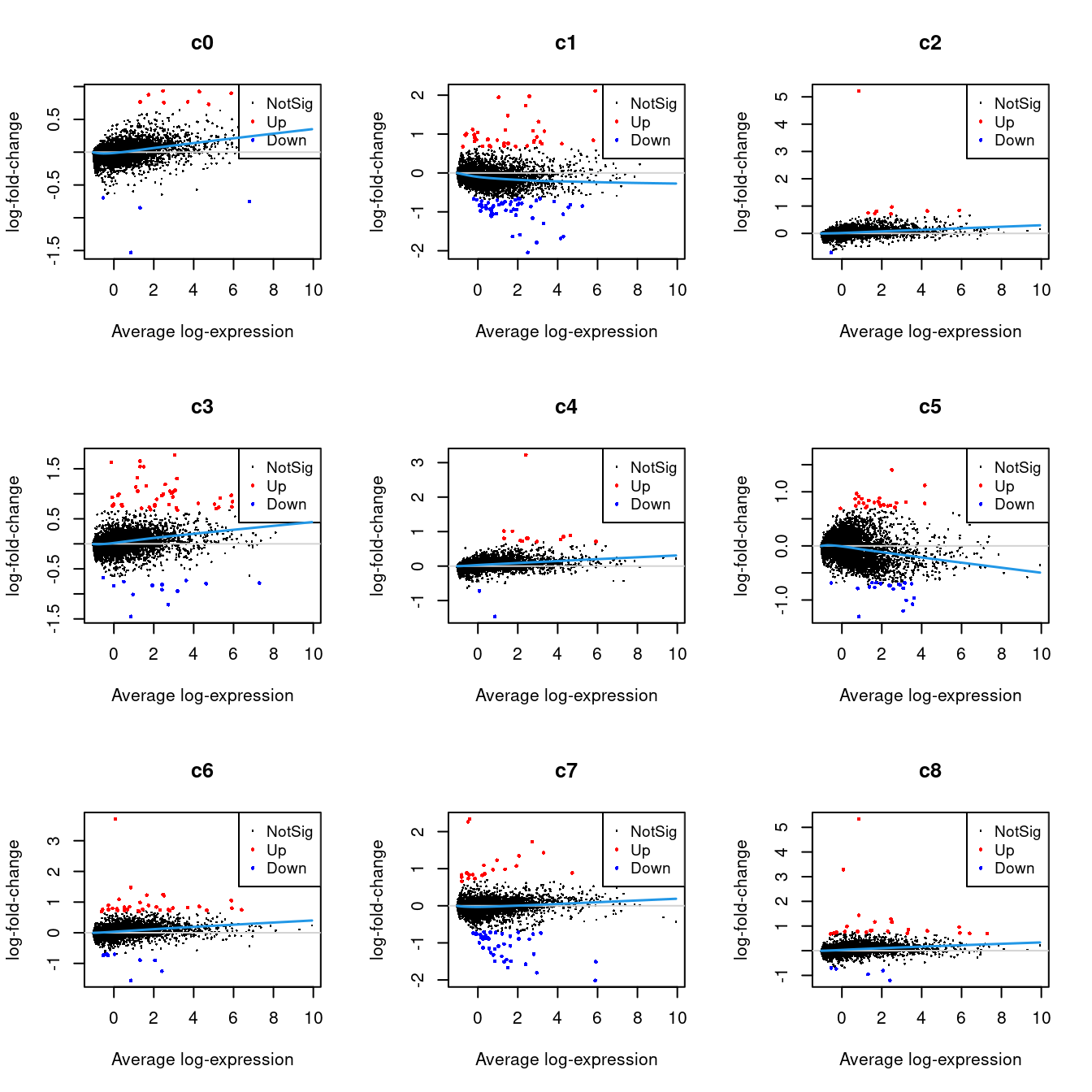
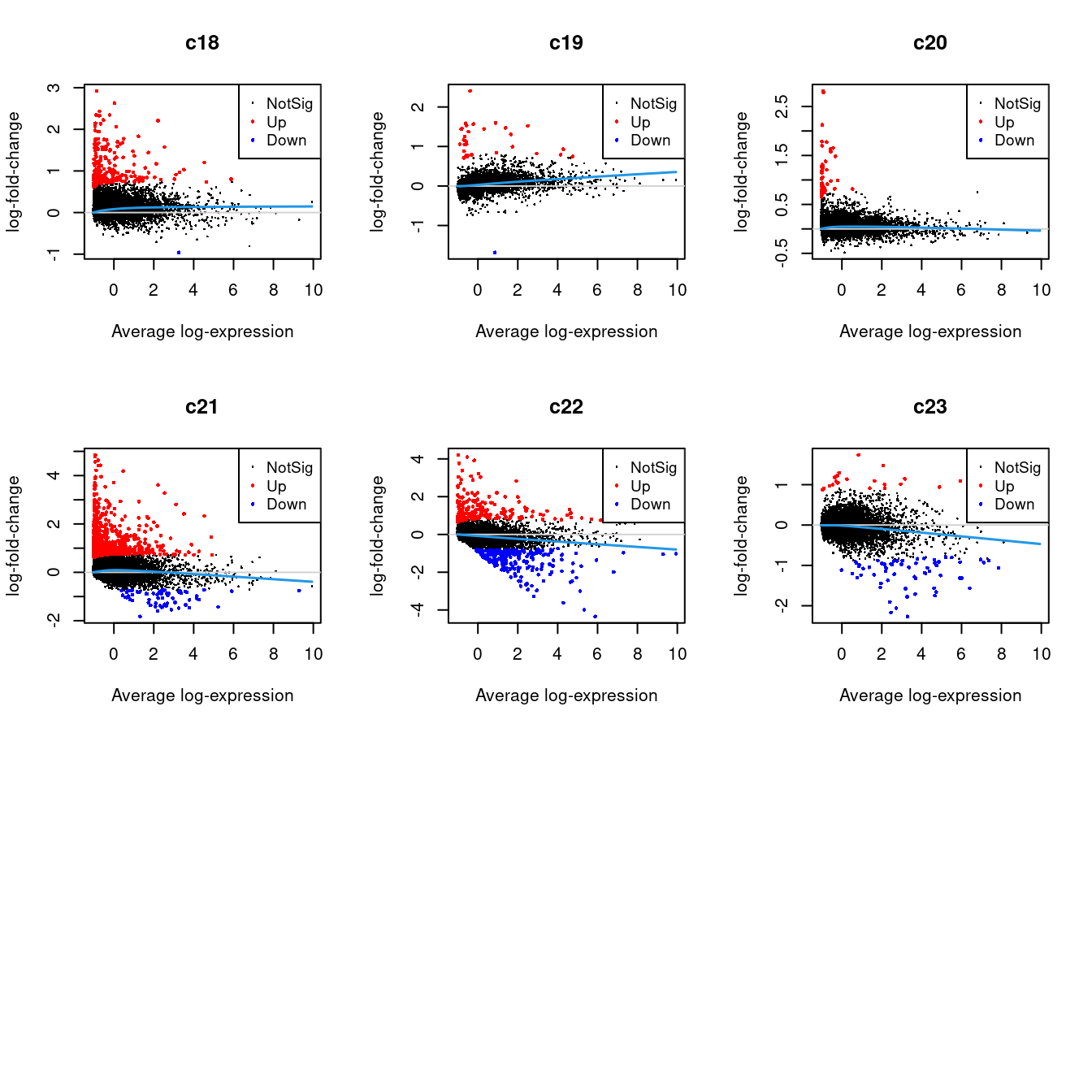
Up 22 153 9 29 75 33 222 30 48 615 223 216.2.1 Mean-difference Plots per Cluster
par(mfrow=c(3,3))
for(i in 1:ncol(mycont)){
plotMD(tr, coef = i, status = dt[,i], hl.cex = 0.5)
abline(h = 0, col = "lightgrey")
lines(lowess(tr$Amean, tr$coefficients[,i]), lwd = 1.5, col = 4)
}
| Version | Author | Date |
|---|---|---|
| f3b7b92 | Jovana Maksimovic | 2022-06-16 |
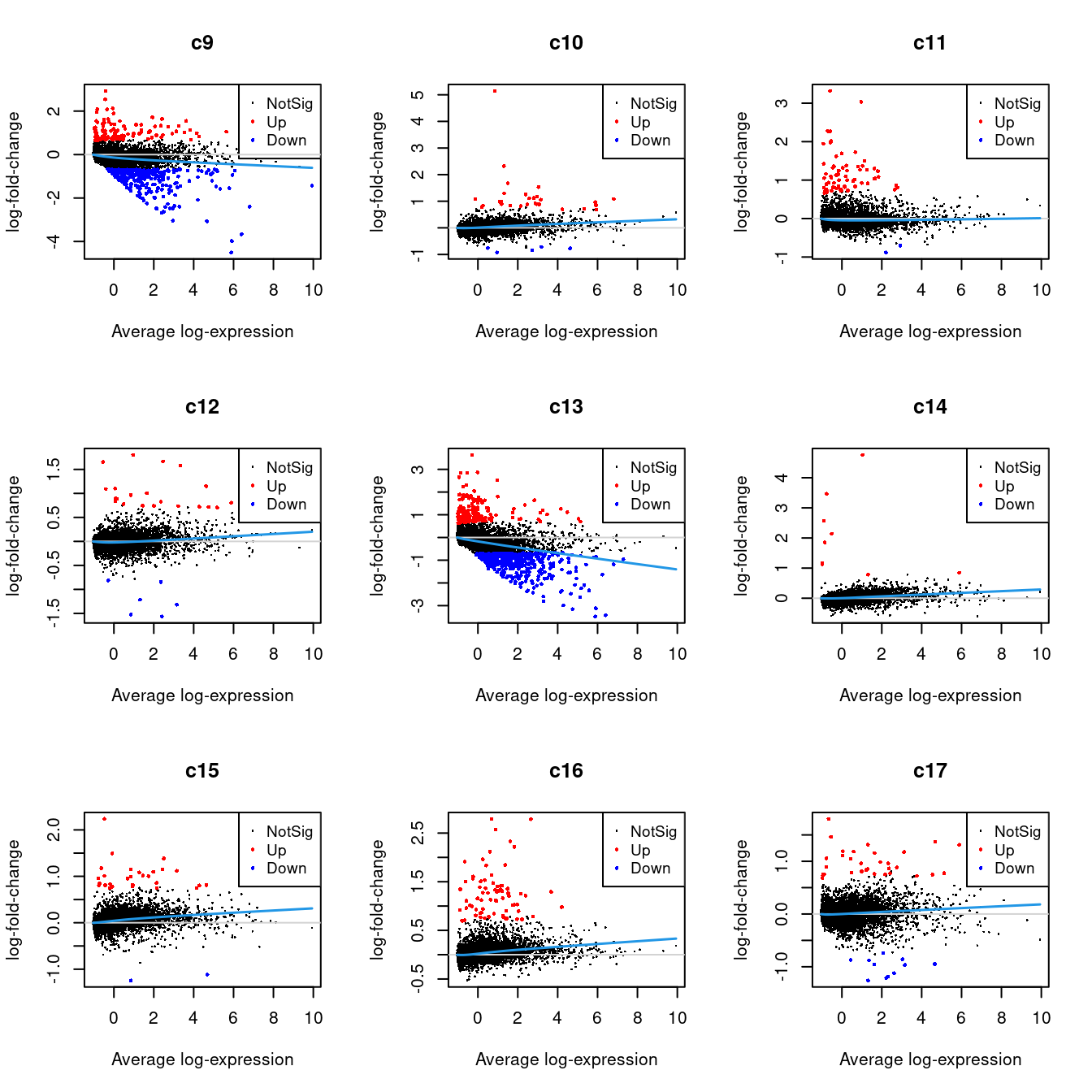
| Version | Author | Date |
|---|---|---|
| f3b7b92 | Jovana Maksimovic | 2022-06-16 |
| Version | Author | Date |
|---|---|---|
| f3b7b92 | Jovana Maksimovic | 2022-06-16 |
6.2.2 Export Marker Genes per cluster
options(scipen=-1, digits = 6)
contnames <- colnames(mycont)
dirName <- here("output/marker-analysis/05-COMBO-macrophages")
if(!dir.exists(dirName)) dir.create(dirName)
getCols <- setNames(c("SYMBOL","ENTREZID"),c("SYMBOL","ENTREZID"))
tr$genes <- data.frame(
lapply(getCols, function(column) {
mapIds(
x = org.Hs.eg.db,
keys = rownames(tr),
keytype = "SYMBOL",
column = column)
}),
row.names = rownames(tr))
gsAnnots <- buildIdx(entrezIDs = tr$genes$ENTREZID, species = "human",
msigdb.gsets = c("c2","c5"))[1] "Loading MSigDB Gene Sets ... "
[1] "Loaded gene sets for the collection c2 ..."
[1] "Indexed the collection c2 ..."
[1] "Created annotation for the collection c2 ..."
[1] "Loaded gene sets for the collection c5 ..."
[1] "Indexed the collection c5 ..."
[1] "Created annotation for the collection c5 ..."
[1] "Building KEGG pathways annotation object ... "reactomeIdx <-gsAnnots$c2@idx[grep("REACTOME",
names(gsAnnots$c2@idx))]
for(i in 1:length(contnames)){
top <- topTreat(tr, coef = i, n = Inf)
top <- top[top$logFC > 0, ]
write.csv(top[1:100, ],
file = glue("{dirName}/up-cluster-{contnames[i]}.csv"))
cameraPR(tr$t[,i], reactomeIdx) %>%
rownames_to_column(var = "Pathway") %>%
slice_head(n = 20) %>%
write_csv(file = here(glue("{dirName}/REACTOME-cluster-{contnames[i]}.csv")))
}6.2.3 Cluster marker gene dot plot
Genes duplicated between clusters are excluded.
sig.genes <- vector("list", ncol(tr))
p <- vector("list",length(sig.genes))
DefaultAssay(seuInt) <- "RNA"
for(i in 1:length(sig.genes)){
top <- topTreat(tr, coef = i, n = Inf)
sig.genes[[i]] <- rownames(top)[top$logFC > 0][1:10]
}
sig <- unlist(sig.genes)
geneCols <- c(rep(rep(c("grey","black"), each = 10), ncol(tr)/2),
rep("grey", 10))[!duplicated(sig)]
DotPlot(seuInt, features = sig[!duplicated(sig)],
group.by = "integrated_snn_res.1",
cols = c("lightgrey", "red"),
dot.scale = 3) +
RotatedAxis() +
FontSize(y.text = 8, x.text = 12) +
labs(y = element_blank(), x = element_blank()) +
coord_flip() +
theme(axis.text.y = element_text(color = geneCols)) +
ggtitle("Top 10 cluster marker genes without duplicates")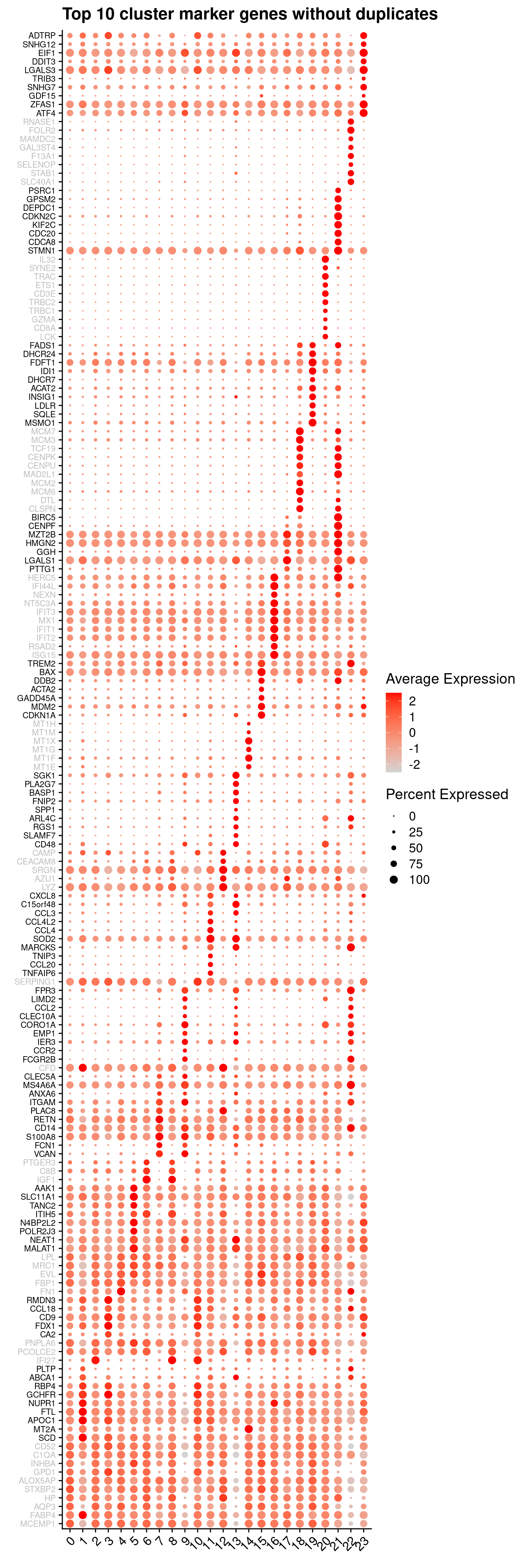
| Version | Author | Date |
|---|---|---|
| f3b7b92 | Jovana Maksimovic | 2022-06-16 |
6.2.4 No. cells per cluster
seuInt@meta.data %>%
ggplot(aes(x = integrated_snn_res.1, fill = integrated_snn_res.1)) +
geom_bar() +
geom_text(aes(label = ..count..), stat = "count",
vjust = -0.5, colour = "black", size = 2) +
theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust = 1)) +
NoLegend()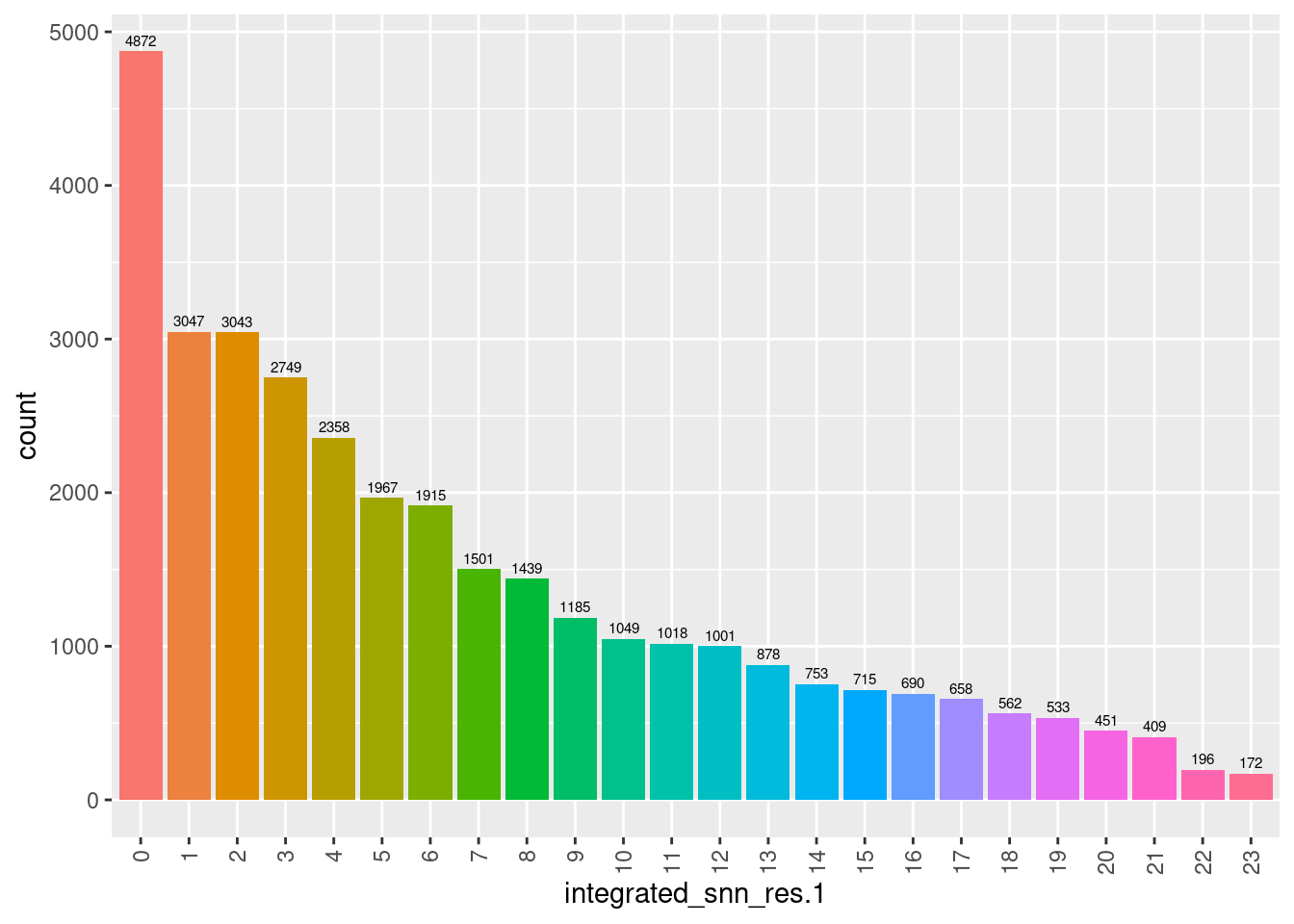
| Version | Author | Date |
|---|---|---|
| f3b7b92 | Jovana Maksimovic | 2022-06-16 |
7 Load protein data
7.1 Add to Seurat object
seuAdt <- readRDS(here("data",
"SCEs",
"05_COMBO.clustered_annotated_adt_diet.SEU.rds"))
seuAdt <- subset(seuAdt, cells = colnames(seuInt))
all(colnames(seuAdt) == colnames(seuInt))[1] TRUEseuInt[["ADT.dsb"]] <- seuAdt[["ADT.dsb"]]
seuInt[["ADT.raw"]] <- seuAdt[["ADT.raw"]]
seuIntAn object of class Seurat
33623 features across 33161 samples within 5 assays
Active assay: RNA (15578 features, 0 variable features)
4 other assays present: SCT, integrated, ADT.dsb, ADT.raw
2 dimensional reductions calculated: pca, umaprm(seuAdt)
gc() used (Mb) gc trigger (Mb) max used (Mb)
Ncells 12082635 645.3 21529594 1149.9 21529594 1149.9
Vcells 2650214870 20219.6 5613736589 42829.5 5613728169 42829.47.2 Load protein annotations
prots <- read.csv(file = here("data",
"sample_sheets",
"TotalSeq-A_Universal_Cocktail_v1.0.csv")) %>%
dplyr::filter(grepl("^A0", id)) %>%
dplyr::filter(!grepl("[Ii]sotype", name)) 7.3 Visualise all ADTs
Normalised with DSB. CITE-seq ADT data was transferred to scRNA-seq using reference mapping and transfer.
cbind(seuInt@meta.data,
as.data.frame(t(seuInt@assays$ADT.dsb@data))) %>%
dplyr::group_by(integrated_snn_res.1, experiment) %>%
dplyr::summarize_at(.vars = prots$id, .funs = median) %>%
pivot_longer(c(-integrated_snn_res.1, -experiment), names_to = "ADT",
values_to = "ADT Exp.") %>%
left_join(prots, by = c("ADT" = "id")) %>%
mutate(Cluster = as.character(integrated_snn_res.1)) %>%
dplyr::rename(Protein = name) |>
dplyr::filter(experiment == 2) |>
ungroup() -> dat
plot(density(dat$`ADT Exp.`))
topMax <- 8
abline(v = topMax, lty = 2, col = "grey")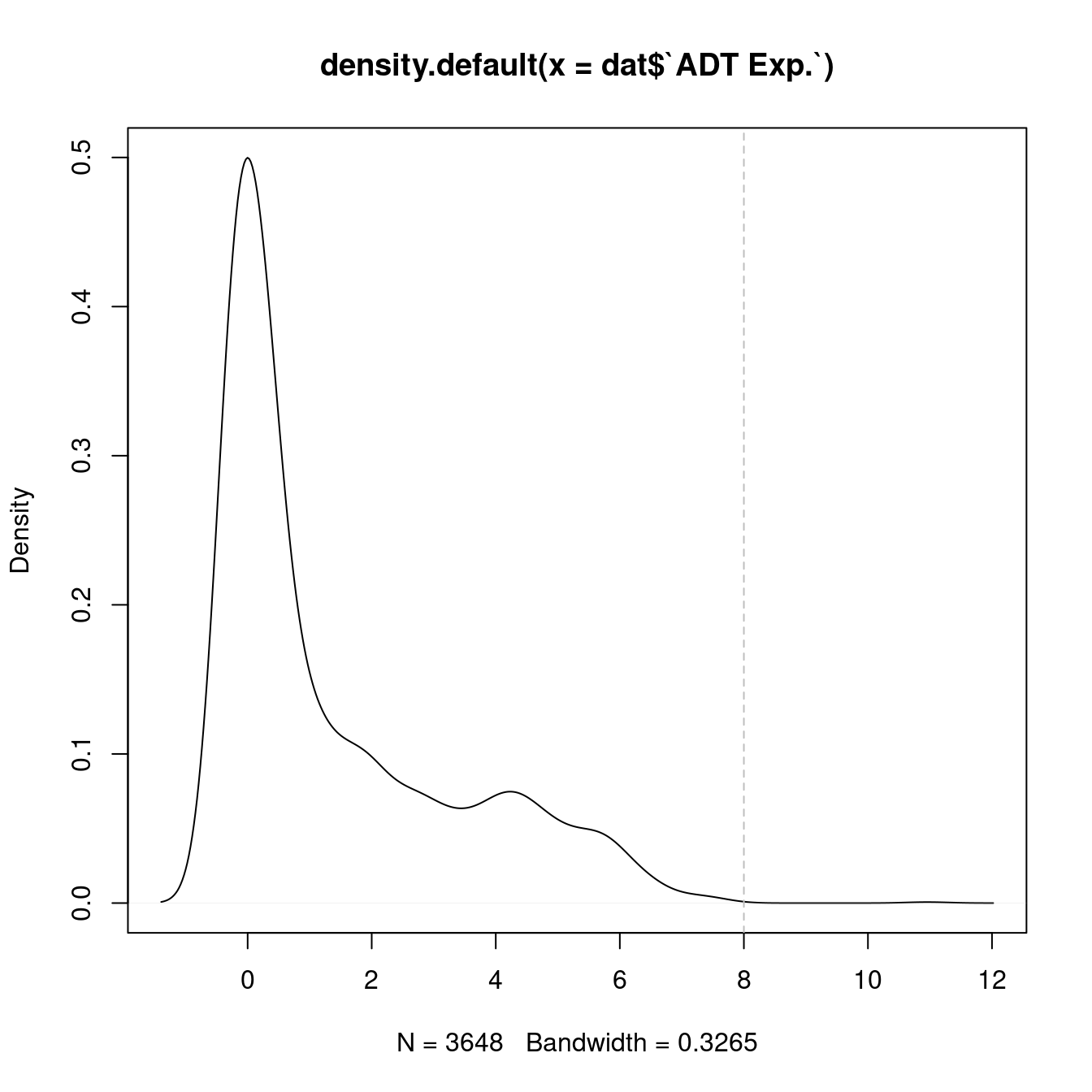
| Version | Author | Date |
|---|---|---|
| f3b7b92 | Jovana Maksimovic | 2022-06-16 |
dat |>
heatmap(
.column = Cluster,
.row = Protein,
.value = `ADT Exp.`,
scale = "none",
rect_gp = grid::gpar(col = "white", lwd = 1),
show_row_names = TRUE,
column_names_gp = grid::gpar(fontsize = 10),
column_title_gp = grid::gpar(fontsize = 12),
row_names_gp = grid::gpar(fontsize = 8),
row_title_gp = grid::gpar(fontsize = 12),
column_title_side = "top",
palette_value = circlize::colorRamp2(seq(-1, topMax, length.out = 256),
viridis::magma(256)),
heatmap_legend_param = list(direction = "vertical")) 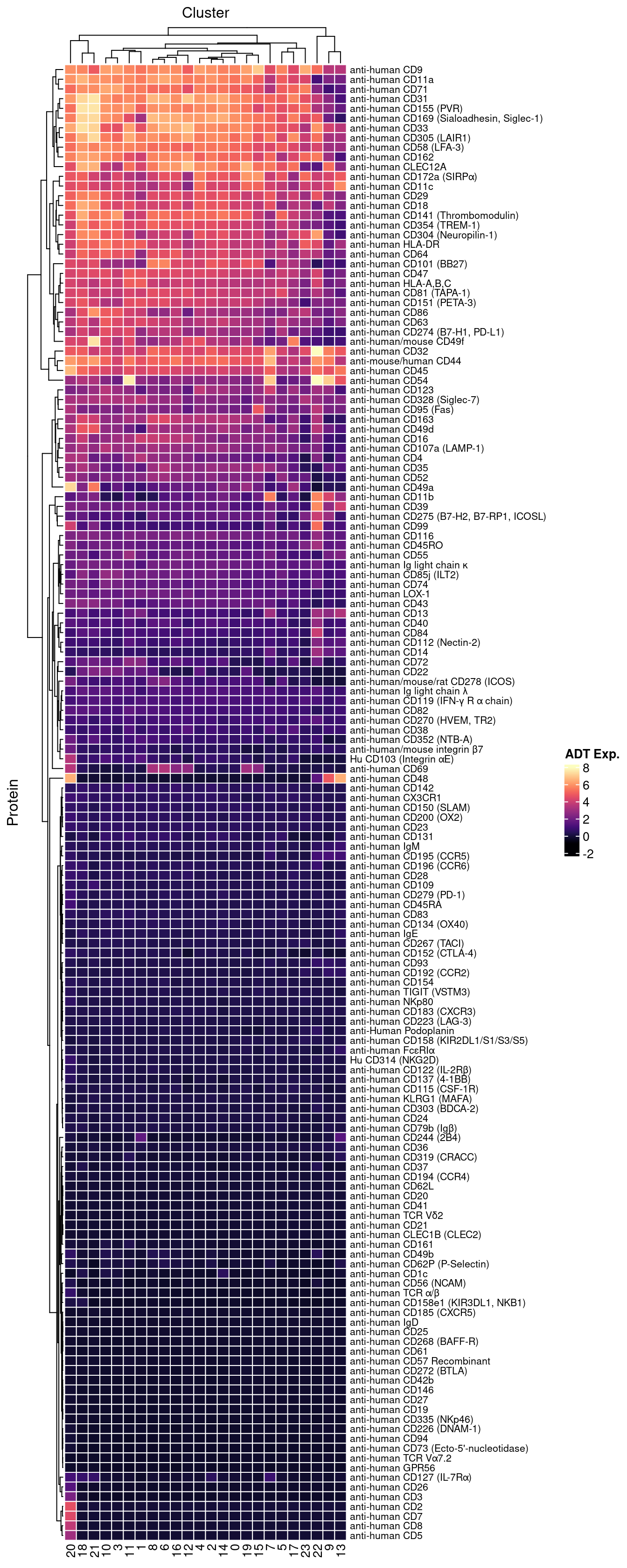
| Version | Author | Date |
|---|---|---|
| f3b7b92 | Jovana Maksimovic | 2022-06-16 |
7.4 Visualise ADTs of interest
adt <- read_csv(file = here("data/Proteins_macs_22.04.22.csv"))
adt <- adt[!duplicated(adt$DNA_ID),]
dat %>%
inner_join(adt, by = c("ADT" = "DNA_ID")) %>%
dplyr::mutate(Protein = `Name for heatmap`) |>
heatmap(
.column = Cluster,
.row = Protein,
.value = `ADT Exp.`,
scale = "none",
rect_gp = grid::gpar(col = "white", lwd = 1),
show_row_names = TRUE,
column_names_gp = grid::gpar(fontsize = 10),
column_title_gp = grid::gpar(fontsize = 12),
row_names_gp = grid::gpar(fontsize = 8),
row_title_gp = grid::gpar(fontsize = 12),
column_title_side = "top",
palette_value = circlize::colorRamp2(seq(-1, topMax, length.out = 256),
viridis::magma(256)),
heatmap_legend_param = list(direction = "vertical")) 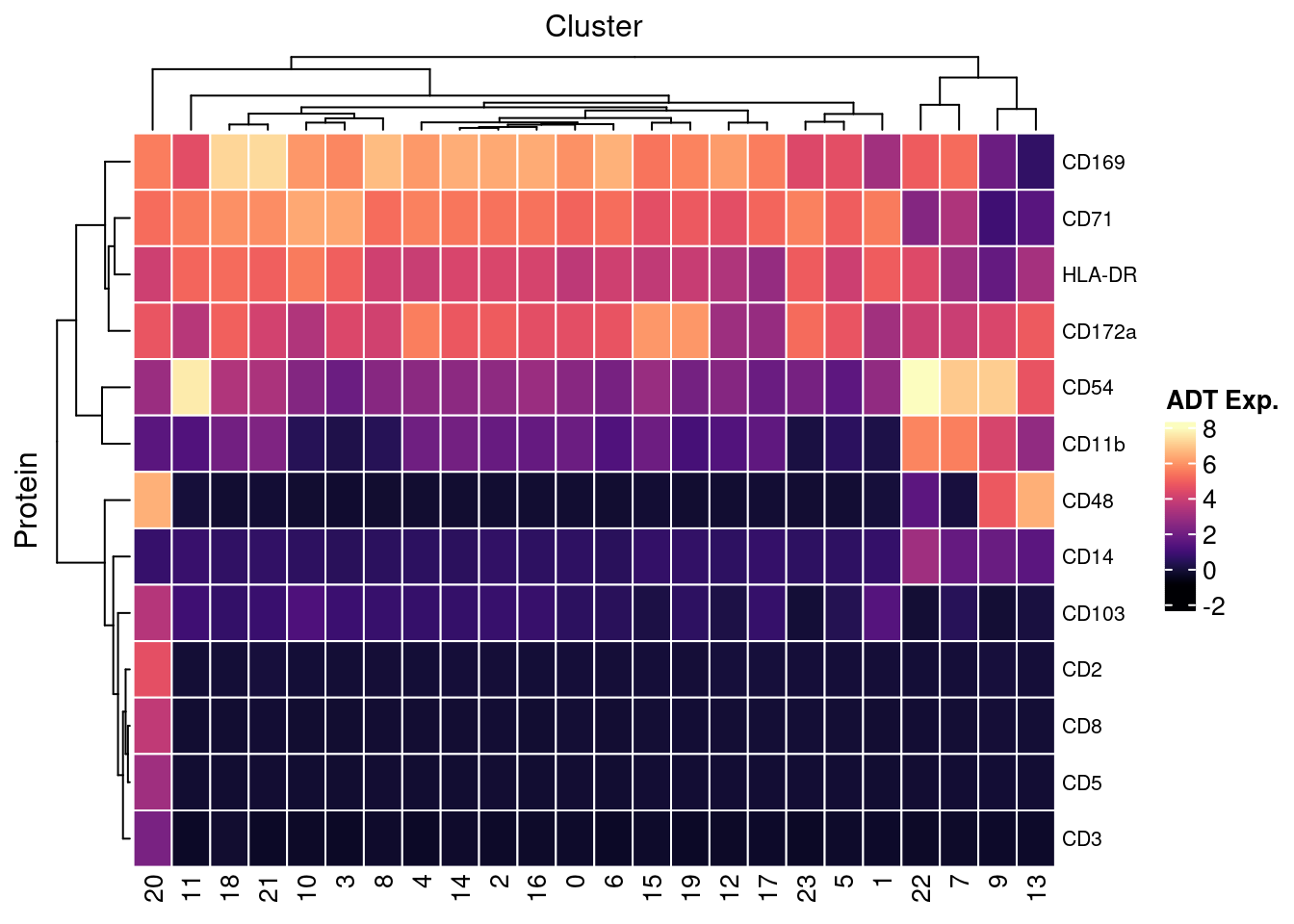
| Version | Author | Date |
|---|---|---|
| f3b7b92 | Jovana Maksimovic | 2022-06-16 |
7.5 Visualise cytokines of interest
markers <- read_csv(file = here("data",
"macrophage_subcluster_cytokines.csv"),
col_names = FALSE)
p <- DotPlot(seuInt,
features = markers$X1,
cols = c("grey", "red"),
dot.scale = 5,
assay = "RNA",
group.by = "integrated_snn_res.1") +
theme(axis.text.x = element_text(angle = 90,
hjust = 1,
vjust = 0.5,
size = 8),
axis.text.y = element_text(size = 8),
text = element_text(size = 8)) +
coord_flip() +
labs(y = "Cluster", x = "Cytokine")
p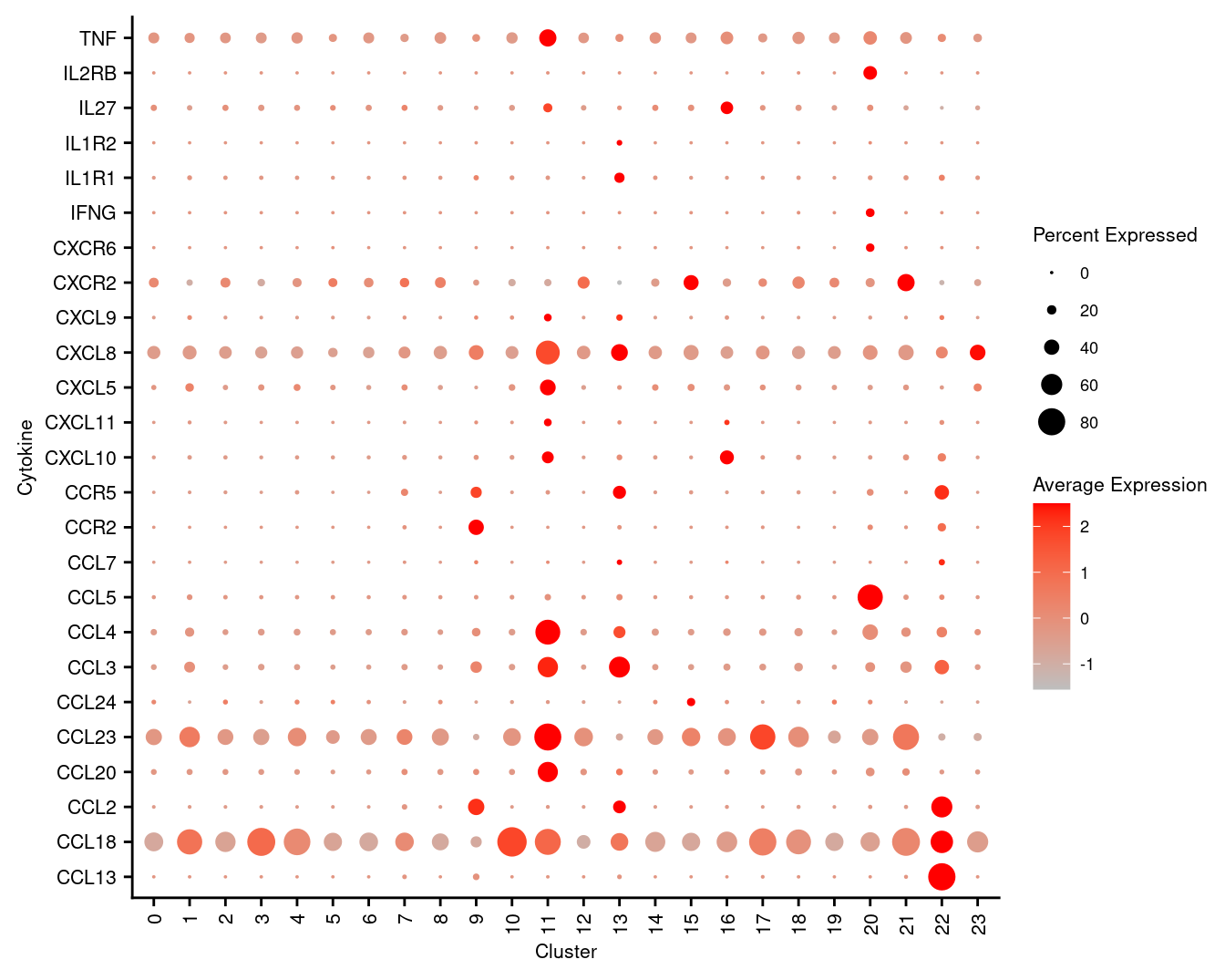
| Version | Author | Date |
|---|---|---|
| f3b7b92 | Jovana Maksimovic | 2022-06-16 |
8 Session info
sessioninfo::session_info()─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.1.0 (2021-05-18)
os CentOS Linux 7 (Core)
system x86_64, linux-gnu
ui X11
language (EN)
collate en_AU.UTF-8
ctype en_AU.UTF-8
tz Australia/Melbourne
date 2022-06-21
pandoc 2.17.1.1 @ /usr/lib/rstudio-server/bin/quarto/bin/ (via rmarkdown)
─ Packages ───────────────────────────────────────────────────────────────────
! package * version date (UTC) lib source
P abind 1.4-5 2016-07-21 [?] CRAN (R 4.1.0)
P annotate 1.72.0 2021-10-26 [?] Bioconductor
P AnnotationDbi * 1.56.2 2021-11-09 [?] Bioconductor
P assertthat 0.2.1 2019-03-21 [?] CRAN (R 4.1.0)
P backports 1.4.1 2021-12-13 [?] CRAN (R 4.1.0)
P beachmat 2.10.0 2021-10-26 [?] Bioconductor
P beeswarm 0.4.0 2021-06-01 [?] CRAN (R 4.1.0)
P Biobase * 2.54.0 2021-10-26 [?] Bioconductor
P BiocGenerics * 0.40.0 2021-10-26 [?] Bioconductor
P BiocManager 1.30.16 2021-06-15 [?] CRAN (R 4.1.0)
P BiocNeighbors 1.12.0 2021-10-26 [?] Bioconductor
P BiocParallel * 1.28.3 2021-12-09 [?] Bioconductor
P BiocSingular 1.10.0 2021-10-26 [?] Bioconductor
P BiocStyle * 2.22.0 2021-10-26 [?] Bioconductor
P Biostrings 2.62.0 2021-10-26 [?] Bioconductor
P bit 4.0.4 2020-08-04 [?] CRAN (R 4.1.0)
P bit64 4.0.5 2020-08-30 [?] CRAN (R 4.0.2)
P bitops 1.0-7 2021-04-24 [?] CRAN (R 4.0.2)
P blob 1.2.2 2021-07-23 [?] CRAN (R 4.1.0)
P bluster 1.4.0 2021-10-26 [?] Bioconductor
P bookdown 0.24 2021-09-02 [?] CRAN (R 4.1.0)
P broom 0.7.11 2022-01-03 [?] CRAN (R 4.1.0)
P bslib 0.3.1 2021-10-06 [?] CRAN (R 4.1.0)
P cachem 1.0.6 2021-08-19 [?] CRAN (R 4.1.0)
P callr 3.7.0 2021-04-20 [?] CRAN (R 4.1.0)
P caTools 1.18.2 2021-03-28 [?] CRAN (R 4.1.0)
P cellranger 1.1.0 2016-07-27 [?] CRAN (R 4.1.0)
P checkmate 2.0.0 2020-02-06 [?] CRAN (R 4.0.2)
P circlize 0.4.13 2021-06-09 [?] CRAN (R 4.1.0)
P cli 3.1.0 2021-10-27 [?] CRAN (R 4.1.0)
P clue 0.3-60 2021-10-11 [?] CRAN (R 4.1.0)
P cluster 2.1.2 2021-04-17 [?] CRAN (R 4.1.0)
P clustree * 0.4.4 2021-11-08 [?] CRAN (R 4.1.0)
P codetools 0.2-18 2020-11-04 [?] CRAN (R 4.1.0)
P colorspace 2.0-2 2021-06-24 [?] CRAN (R 4.0.2)
P ComplexHeatmap 2.10.0 2021-10-26 [?] Bioconductor
P cowplot 1.1.1 2020-12-30 [?] CRAN (R 4.0.2)
P crayon 1.4.2 2021-10-29 [?] CRAN (R 4.1.0)
P data.table 1.14.2 2021-09-27 [?] CRAN (R 4.1.0)
P DBI 1.1.2 2021-12-20 [?] CRAN (R 4.1.0)
P dbplyr 2.1.1 2021-04-06 [?] CRAN (R 4.1.0)
P DelayedArray 0.20.0 2021-10-26 [?] Bioconductor
P DelayedMatrixStats 1.16.0 2021-10-26 [?] Bioconductor
P deldir 1.0-6 2021-10-23 [?] CRAN (R 4.1.0)
P dendextend 1.15.2 2021-10-28 [?] CRAN (R 4.1.0)
P digest 0.6.29 2021-12-01 [?] CRAN (R 4.1.0)
P doParallel 1.0.16 2020-10-16 [?] CRAN (R 4.0.2)
P doRNG 1.8.2 2020-01-27 [?] CRAN (R 4.1.0)
P dplyr * 1.0.7 2021-06-18 [?] CRAN (R 4.1.0)
P dqrng 0.3.0 2021-05-01 [?] CRAN (R 4.1.0)
P DropletUtils * 1.14.1 2021-11-08 [?] Bioconductor
P DT 0.20 2021-11-15 [?] CRAN (R 4.1.0)
P edgeR * 3.36.0 2021-10-26 [?] Bioconductor
P EGSEA * 1.22.0 2021-10-26 [?] Bioconductor
P EGSEAdata 1.22.0 2021-10-30 [?] Bioconductor
P ellipsis 0.3.2 2021-04-29 [?] CRAN (R 4.0.2)
P evaluate 0.14 2019-05-28 [?] CRAN (R 4.0.2)
P fansi 1.0.0 2022-01-10 [?] CRAN (R 4.1.0)
P farver 2.1.0 2021-02-28 [?] CRAN (R 4.0.2)
P fastmap 1.1.0 2021-01-25 [?] CRAN (R 4.1.0)
P fitdistrplus 1.1-6 2021-09-28 [?] CRAN (R 4.1.0)
P forcats * 0.5.1 2021-01-27 [?] CRAN (R 4.1.0)
P foreach 1.5.1 2020-10-15 [?] CRAN (R 4.0.2)
P fs 1.5.2 2021-12-08 [?] CRAN (R 4.1.0)
P future 1.23.0 2021-10-31 [?] CRAN (R 4.1.0)
P future.apply 1.8.1 2021-08-10 [?] CRAN (R 4.1.0)
P gage * 2.44.0 2021-10-26 [?] Bioconductor
P generics 0.1.1 2021-10-25 [?] CRAN (R 4.1.0)
GenomeInfoDb * 1.30.1 2022-01-30 [1] Bioconductor
P GenomeInfoDbData 1.2.7 2021-12-21 [?] Bioconductor
P GenomicRanges * 1.46.1 2021-11-18 [?] Bioconductor
P GetoptLong 1.0.5 2020-12-15 [?] CRAN (R 4.0.2)
P getPass 0.2-2 2017-07-21 [?] CRAN (R 4.0.2)
P ggbeeswarm 0.6.0 2017-08-07 [?] CRAN (R 4.1.0)
P ggforce 0.3.3 2021-03-05 [?] CRAN (R 4.1.0)
P ggplot2 * 3.3.5 2021-06-25 [?] CRAN (R 4.0.2)
P ggraph * 2.0.5 2021-02-23 [?] CRAN (R 4.1.0)
P ggrepel 0.9.1 2021-01-15 [?] CRAN (R 4.1.0)
P ggridges 0.5.3 2021-01-08 [?] CRAN (R 4.1.0)
P git2r 0.29.0 2021-11-22 [?] CRAN (R 4.1.0)
P glmGamPoi * 1.6.0 2021-10-26 [?] Bioconductor
P GlobalOptions 0.1.2 2020-06-10 [?] CRAN (R 4.1.0)
P globals 0.14.0 2020-11-22 [?] CRAN (R 4.0.2)
P globaltest 5.48.0 2021-10-26 [?] Bioconductor
P glue * 1.6.0 2021-12-17 [?] CRAN (R 4.1.0)
P GO.db * 3.14.0 2021-12-21 [?] Bioconductor
P goftest 1.2-3 2021-10-07 [?] CRAN (R 4.1.0)
P gplots 3.1.1 2020-11-28 [?] CRAN (R 4.0.2)
P graph * 1.72.0 2021-10-26 [?] Bioconductor
P graphlayouts 0.8.0 2022-01-03 [?] CRAN (R 4.1.0)
P gridExtra 2.3 2017-09-09 [?] CRAN (R 4.1.0)
P GSA 1.03.1 2019-01-31 [?] CRAN (R 4.1.0)
P GSEABase 1.56.0 2021-10-26 [?] Bioconductor
P GSVA 1.42.0 2021-10-26 [?] Bioconductor
P gtable 0.3.0 2019-03-25 [?] CRAN (R 4.1.0)
P gtools 3.9.2 2021-06-06 [?] CRAN (R 4.1.0)
P haven 2.4.3 2021-08-04 [?] CRAN (R 4.1.0)
P HDF5Array 1.22.1 2021-11-14 [?] Bioconductor
P here * 1.0.1 2020-12-13 [?] CRAN (R 4.0.2)
P hgu133a.db 3.13.0 2022-01-24 [?] Bioconductor
P hgu133plus2.db 3.13.0 2022-01-24 [?] Bioconductor
P highr 0.9 2021-04-16 [?] CRAN (R 4.1.0)
P hms 1.1.1 2021-09-26 [?] CRAN (R 4.1.0)
P htmltools 0.5.2 2021-08-25 [?] CRAN (R 4.1.0)
P HTMLUtils 0.1.7 2015-01-17 [?] CRAN (R 4.1.0)
P htmlwidgets 1.5.4 2021-09-08 [?] CRAN (R 4.1.0)
P httpuv 1.6.5 2022-01-05 [?] CRAN (R 4.1.0)
P httr 1.4.2 2020-07-20 [?] CRAN (R 4.1.0)
P hwriter 1.3.2 2014-09-10 [?] CRAN (R 4.1.0)
P ica 1.0-2 2018-05-24 [?] CRAN (R 4.1.0)
P igraph 1.2.11 2022-01-04 [?] CRAN (R 4.1.0)
P IRanges * 2.28.0 2021-10-26 [?] Bioconductor
P irlba 2.3.5 2021-12-06 [?] CRAN (R 4.1.0)
P iterators 1.0.13 2020-10-15 [?] CRAN (R 4.0.2)
P jquerylib 0.1.4 2021-04-26 [?] CRAN (R 4.1.0)
P jsonlite 1.7.2 2020-12-09 [?] CRAN (R 4.0.2)
P KEGGdzPathwaysGEO 1.32.0 2021-10-30 [?] Bioconductor
P KEGGgraph 1.54.0 2021-10-26 [?] Bioconductor
P KEGGREST 1.34.0 2021-10-26 [?] Bioconductor
P KernSmooth 2.23-20 2021-05-03 [?] CRAN (R 4.1.0)
P knitr 1.37 2021-12-16 [?] CRAN (R 4.1.0)
P labeling 0.4.2 2020-10-20 [?] CRAN (R 4.0.2)
P later 1.3.0 2021-08-18 [?] CRAN (R 4.1.0)
P lattice 0.20-45 2021-09-22 [?] CRAN (R 4.1.0)
P lazyeval 0.2.2 2019-03-15 [?] CRAN (R 4.1.0)
P leiden 0.3.9 2021-07-27 [?] CRAN (R 4.1.0)
P lifecycle 1.0.1 2021-09-24 [?] CRAN (R 4.1.0)
P limma * 3.50.0 2021-10-26 [?] Bioconductor
P listenv 0.8.0 2019-12-05 [?] CRAN (R 4.1.0)
P lmtest 0.9-39 2021-11-07 [?] CRAN (R 4.1.0)
P locfit 1.5-9.4 2020-03-25 [?] CRAN (R 4.1.0)
P lubridate 1.8.0 2021-10-07 [?] CRAN (R 4.1.0)
P magrittr 2.0.1 2020-11-17 [?] CRAN (R 4.0.2)
P MASS 7.3-53.1 2021-02-12 [?] CRAN (R 4.0.2)
P mathjaxr 1.4-0 2021-03-01 [?] CRAN (R 4.1.0)
P Matrix 1.4-0 2021-12-08 [?] CRAN (R 4.1.0)
P MatrixGenerics * 1.6.0 2021-10-26 [?] Bioconductor
P matrixStats * 0.61.0 2021-09-17 [?] CRAN (R 4.1.0)
P memoise 2.0.1 2021-11-26 [?] CRAN (R 4.1.0)
P metap 1.7 2021-12-16 [?] CRAN (R 4.1.0)
P metapod 1.2.0 2021-10-26 [?] Bioconductor
P mgcv 1.8-38 2021-10-06 [?] CRAN (R 4.1.0)
P mime 0.12 2021-09-28 [?] CRAN (R 4.1.0)
P miniUI 0.1.1.1 2018-05-18 [?] CRAN (R 4.1.0)
P mnormt 2.0.2 2020-09-01 [?] CRAN (R 4.0.2)
P modelr 0.1.8 2020-05-19 [?] CRAN (R 4.0.2)
P multcomp 1.4-18 2022-01-04 [?] CRAN (R 4.1.0)
P multtest 2.50.0 2021-10-26 [?] Bioconductor
P munsell 0.5.0 2018-06-12 [?] CRAN (R 4.1.0)
P mutoss 0.1-12 2017-12-04 [?] CRAN (R 4.1.0)
P mvtnorm 1.1-3 2021-10-08 [?] CRAN (R 4.1.0)
P nlme 3.1-153 2021-09-07 [?] CRAN (R 4.1.0)
P numDeriv 2016.8-1.1 2019-06-06 [?] CRAN (R 4.1.0)
P org.Hs.eg.db * 3.14.0 2021-12-21 [?] Bioconductor
P org.Mm.eg.db 3.14.0 2022-01-24 [?] Bioconductor
P org.Rn.eg.db 3.14.0 2022-01-24 [?] Bioconductor
P PADOG 1.36.0 2021-10-26 [?] Bioconductor
P paletteer * 1.4.0 2021-07-20 [?] CRAN (R 4.1.0)
P parallelly 1.30.0 2021-12-17 [?] CRAN (R 4.1.0)
P patchwork * 1.1.1 2020-12-17 [?] CRAN (R 4.0.2)
P pathview * 1.34.0 2021-10-26 [?] Bioconductor
P pbapply 1.5-0 2021-09-16 [?] CRAN (R 4.1.0)
P pillar 1.6.4 2021-10-18 [?] CRAN (R 4.1.0)
P pkgconfig 2.0.3 2019-09-22 [?] CRAN (R 4.1.0)
P plotly 4.10.0 2021-10-09 [?] CRAN (R 4.1.0)
P plotrix 3.8-2 2021-09-08 [?] CRAN (R 4.1.0)
P plyr 1.8.6 2020-03-03 [?] CRAN (R 4.0.2)
P png 0.1-7 2013-12-03 [?] CRAN (R 4.1.0)
P polyclip 1.10-0 2019-03-14 [?] CRAN (R 4.1.0)
P processx 3.5.2 2021-04-30 [?] CRAN (R 4.1.0)
P promises 1.2.0.1 2021-02-11 [?] CRAN (R 4.0.2)
P ps 1.6.0 2021-02-28 [?] CRAN (R 4.1.0)
P purrr * 0.3.4 2020-04-17 [?] CRAN (R 4.0.2)
P R.methodsS3 1.8.1 2020-08-26 [?] CRAN (R 4.0.2)
P R.oo 1.24.0 2020-08-26 [?] CRAN (R 4.0.2)
P R.utils 2.11.0 2021-09-26 [?] CRAN (R 4.1.0)
P R2HTML 2.3.2 2016-06-23 [?] CRAN (R 4.1.0)
P R6 2.5.1 2021-08-19 [?] CRAN (R 4.1.0)
P RANN 2.6.1 2019-01-08 [?] CRAN (R 4.1.0)
P rbibutils 2.2.7 2021-12-07 [?] CRAN (R 4.1.0)
P RColorBrewer 1.1-2 2014-12-07 [?] CRAN (R 4.0.2)
P Rcpp 1.0.7 2021-07-07 [?] CRAN (R 4.1.0)
P RcppAnnoy 0.0.19 2021-07-30 [?] CRAN (R 4.1.0)
RCurl 1.98-1.6 2022-02-08 [1] CRAN (R 4.1.0)
P Rdpack 2.1.3 2021-12-08 [?] CRAN (R 4.1.0)
P readr * 2.1.1 2021-11-30 [?] CRAN (R 4.1.0)
P readxl 1.3.1 2019-03-13 [?] CRAN (R 4.1.0)
P rematch2 2.1.2 2020-05-01 [?] CRAN (R 4.1.0)
P renv 0.15.0-14 2022-01-10 [?] Github (rstudio/renv@a3b90eb)
P reprex 2.0.1 2021-08-05 [?] CRAN (R 4.1.0)
P reshape2 1.4.4 2020-04-09 [?] CRAN (R 4.1.0)
P reticulate 1.22 2021-09-17 [?] CRAN (R 4.1.0)
P Rgraphviz 2.38.0 2021-10-26 [?] Bioconductor
P rhdf5 2.38.0 2021-10-26 [?] Bioconductor
P rhdf5filters 1.6.0 2021-10-26 [?] Bioconductor
P Rhdf5lib 1.16.0 2021-10-26 [?] Bioconductor
P rjson 0.2.21 2022-01-09 [?] CRAN (R 4.1.0)
P rlang 0.4.12 2021-10-18 [?] CRAN (R 4.1.0)
P rmarkdown 2.11 2021-09-14 [?] CRAN (R 4.1.0)
P rngtools 1.5.2 2021-09-20 [?] CRAN (R 4.1.0)
P ROCR 1.0-11 2020-05-02 [?] CRAN (R 4.1.0)
P rpart 4.1-15 2019-04-12 [?] CRAN (R 4.1.0)
P rprojroot 2.0.2 2020-11-15 [?] CRAN (R 4.0.2)
P RSpectra 0.16-0 2019-12-01 [?] CRAN (R 4.1.0)
P RSQLite 2.2.9 2021-12-06 [?] CRAN (R 4.1.0)
P rstudioapi 0.13 2020-11-12 [?] CRAN (R 4.0.2)
P rsvd 1.0.5 2021-04-16 [?] CRAN (R 4.1.0)
P Rtsne 0.15 2018-11-10 [?] CRAN (R 4.1.0)
P rvest 1.0.2 2021-10-16 [?] CRAN (R 4.1.0)
P S4Vectors * 0.32.3 2021-11-21 [?] Bioconductor
P safe 3.34.0 2021-10-26 [?] Bioconductor
P sandwich 3.0-1 2021-05-18 [?] CRAN (R 4.1.0)
P sass 0.4.0 2021-05-12 [?] CRAN (R 4.1.0)
P ScaledMatrix 1.2.0 2021-10-26 [?] Bioconductor
P scales 1.1.1 2020-05-11 [?] CRAN (R 4.0.2)
P scater * 1.22.0 2021-10-26 [?] Bioconductor
P scattermore 0.7 2020-11-24 [?] CRAN (R 4.1.0)
P scran * 1.22.1 2021-11-14 [?] Bioconductor
P sctransform 0.3.3 2022-01-13 [?] CRAN (R 4.1.0)
P scuttle * 1.4.0 2021-10-26 [?] Bioconductor
P sessioninfo 1.2.2 2021-12-06 [?] CRAN (R 4.1.0)
P Seurat * 4.0.6 2021-12-16 [?] CRAN (R 4.1.0)
P SeuratObject * 4.0.4 2021-11-23 [?] CRAN (R 4.1.0)
P shape 1.4.6 2021-05-19 [?] CRAN (R 4.1.0)
P shiny 1.7.1 2021-10-02 [?] CRAN (R 4.1.0)
P SingleCellExperiment * 1.16.0 2021-10-26 [?] Bioconductor
P sn 2.0.1 2021-11-26 [?] CRAN (R 4.1.0)
P SparseM * 1.81 2021-02-18 [?] CRAN (R 4.1.0)
P sparseMatrixStats 1.6.0 2021-10-26 [?] Bioconductor
P spatstat.core 2.3-2 2021-11-26 [?] CRAN (R 4.1.0)
P spatstat.data 2.1-2 2021-12-17 [?] CRAN (R 4.1.0)
P spatstat.geom 2.3-1 2021-12-10 [?] CRAN (R 4.1.0)
P spatstat.sparse 2.1-0 2021-12-17 [?] CRAN (R 4.1.0)
P spatstat.utils 2.3-0 2021-12-12 [?] CRAN (R 4.1.0)
P statmod 1.4.36 2021-05-10 [?] CRAN (R 4.1.0)
P stringi 1.7.6 2021-11-29 [?] CRAN (R 4.1.0)
P stringr * 1.4.0 2019-02-10 [?] CRAN (R 4.0.2)
P SummarizedExperiment * 1.24.0 2021-10-26 [?] Bioconductor
P survival 3.2-13 2021-08-24 [?] CRAN (R 4.1.0)
P tensor 1.5 2012-05-05 [?] CRAN (R 4.1.0)
P TFisher 0.2.0 2018-03-21 [?] CRAN (R 4.1.0)
P TH.data 1.1-0 2021-09-27 [?] CRAN (R 4.1.0)
P tibble * 3.1.6 2021-11-07 [?] CRAN (R 4.1.0)
P tidygraph 1.2.0 2020-05-12 [?] CRAN (R 4.0.2)
P tidyHeatmap * 1.7.0 2022-05-13 [?] Github (stemangiola/tidyHeatmap@241aec2)
P tidyr * 1.1.4 2021-09-27 [?] CRAN (R 4.1.0)
P tidyselect 1.1.1 2021-04-30 [?] CRAN (R 4.1.0)
P tidyverse * 1.3.1 2021-04-15 [?] CRAN (R 4.1.0)
P tmvnsim 1.0-2 2016-12-15 [?] CRAN (R 4.1.0)
P topGO * 2.46.0 2021-10-26 [?] Bioconductor
P tweenr 1.0.2 2021-03-23 [?] CRAN (R 4.1.0)
P tzdb 0.2.0 2021-10-27 [?] CRAN (R 4.1.0)
P utf8 1.2.2 2021-07-24 [?] CRAN (R 4.1.0)
P uwot 0.1.11 2021-12-02 [?] CRAN (R 4.1.0)
P vctrs 0.3.8 2021-04-29 [?] CRAN (R 4.0.2)
P vipor 0.4.5 2017-03-22 [?] CRAN (R 4.1.0)
P viridis 0.6.2 2021-10-13 [?] CRAN (R 4.1.0)
P viridisLite 0.4.0 2021-04-13 [?] CRAN (R 4.0.2)
P vroom 1.5.7 2021-11-30 [?] CRAN (R 4.1.0)
P whisker 0.4 2019-08-28 [?] CRAN (R 4.0.2)
P withr 2.4.3 2021-11-30 [?] CRAN (R 4.1.0)
P workflowr * 1.7.0 2021-12-21 [?] CRAN (R 4.1.0)
P xfun 0.29 2021-12-14 [?] CRAN (R 4.1.0)
P XML 3.99-0.8 2021-09-17 [?] CRAN (R 4.1.0)
P xml2 1.3.3 2021-11-30 [?] CRAN (R 4.1.0)
P xtable 1.8-4 2019-04-21 [?] CRAN (R 4.1.0)
P XVector 0.34.0 2021-10-26 [?] Bioconductor
P yaml 2.2.1 2020-02-01 [?] CRAN (R 4.0.2)
P zlibbioc 1.40.0 2021-10-26 [?] Bioconductor
P zoo 1.8-9 2021-03-09 [?] CRAN (R 4.1.0)
[1] /oshlack_lab/jovana.maksimovic/projects/MCRI/melanie.neeland/paed-cf-cite-seq/renv/library/R-4.1/x86_64-pc-linux-gnu
[2] /config/binaries/R/4.1.0/lib64/R/library
P ── Loaded and on-disk path mismatch.
──────────────────────────────────────────────────────────────────────────────9 References
sessionInfo()R version 4.1.0 (2021-05-18)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: CentOS Linux 7 (Core)
Matrix products: default
BLAS: /config/binaries/R/4.1.0/lib64/R/lib/libRblas.so
LAPACK: /config/binaries/R/4.1.0/lib64/R/lib/libRlapack.so
locale:
[1] LC_CTYPE=en_AU.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_AU.UTF-8 LC_COLLATE=en_AU.UTF-8
[5] LC_MONETARY=en_AU.UTF-8 LC_MESSAGES=en_AU.UTF-8
[7] LC_PAPER=en_AU.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_AU.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats4 stats graphics grDevices datasets utils methods
[8] base
other attached packages:
[1] EGSEA_1.22.0 pathview_1.34.0
[3] topGO_2.46.0 SparseM_1.81
[5] GO.db_3.14.0 graph_1.72.0
[7] gage_2.44.0 org.Hs.eg.db_3.14.0
[9] AnnotationDbi_1.56.2 edgeR_3.36.0
[11] limma_3.50.0 tidyHeatmap_1.7.0
[13] paletteer_1.4.0 BiocParallel_1.28.3
[15] glmGamPoi_1.6.0 clustree_0.4.4
[17] ggraph_2.0.5 patchwork_1.1.1
[19] SeuratObject_4.0.4 Seurat_4.0.6
[21] scater_1.22.0 scran_1.22.1
[23] scuttle_1.4.0 DropletUtils_1.14.1
[25] SingleCellExperiment_1.16.0 SummarizedExperiment_1.24.0
[27] Biobase_2.54.0 GenomicRanges_1.46.1
[29] GenomeInfoDb_1.30.1 IRanges_2.28.0
[31] S4Vectors_0.32.3 BiocGenerics_0.40.0
[33] MatrixGenerics_1.6.0 matrixStats_0.61.0
[35] glue_1.6.0 here_1.0.1
[37] forcats_0.5.1 stringr_1.4.0
[39] dplyr_1.0.7 purrr_0.3.4
[41] readr_2.1.1 tidyr_1.1.4
[43] tibble_3.1.6 ggplot2_3.3.5
[45] tidyverse_1.3.1 BiocStyle_2.22.0
[47] workflowr_1.7.0
loaded via a namespace (and not attached):
[1] rsvd_1.0.5 ica_1.0-2
[3] ps_1.6.0 foreach_1.5.1
[5] lmtest_0.9-39 rprojroot_2.0.2
[7] crayon_1.4.2 rbibutils_2.2.7
[9] spatstat.core_2.3-2 MASS_7.3-53.1
[11] rhdf5filters_1.6.0 nlme_3.1-153
[13] backports_1.4.1 reprex_2.0.1
[15] rlang_0.4.12 XVector_0.34.0
[17] ROCR_1.0-11 readxl_1.3.1
[19] irlba_2.3.5 callr_3.7.0
[21] rjson_0.2.21 globaltest_5.48.0
[23] bit64_4.0.5 rngtools_1.5.2
[25] sctransform_0.3.3 parallel_4.1.0
[27] processx_3.5.2 vipor_0.4.5
[29] spatstat.sparse_2.1-0 R2HTML_2.3.2
[31] spatstat.geom_2.3-1 haven_2.4.3
[33] tidyselect_1.1.1 fitdistrplus_1.1-6
[35] XML_3.99-0.8 zoo_1.8-9
[37] org.Mm.eg.db_3.14.0 xtable_1.8-4
[39] magrittr_2.0.1 evaluate_0.14
[41] Rdpack_2.1.3 cli_3.1.0
[43] zlibbioc_1.40.0 sn_2.0.1
[45] hwriter_1.3.2 doRNG_1.8.2
[47] rstudioapi_0.13 miniUI_0.1.1.1
[49] whisker_0.4 bslib_0.3.1
[51] rpart_4.1-15 mathjaxr_1.4-0
[53] GSA_1.03.1 KEGGdzPathwaysGEO_1.32.0
[55] shiny_1.7.1 GSVA_1.42.0
[57] BiocSingular_1.10.0 xfun_0.29
[59] clue_0.3-60 org.Rn.eg.db_3.14.0
[61] multtest_2.50.0 cluster_2.1.2
[63] caTools_1.18.2 tidygraph_1.2.0
[65] KEGGREST_1.34.0 ggrepel_0.9.1
[67] listenv_0.8.0 dendextend_1.15.2
[69] Biostrings_2.62.0 png_0.1-7
[71] future_1.23.0 withr_2.4.3
[73] bitops_1.0-7 ggforce_0.3.3
[75] plyr_1.8.6 cellranger_1.1.0
[77] PADOG_1.36.0 GSEABase_1.56.0
[79] dqrng_0.3.0 pillar_1.6.4
[81] gplots_3.1.1 GlobalOptions_0.1.2
[83] cachem_1.0.6 multcomp_1.4-18
[85] fs_1.5.2 GetoptLong_1.0.5
[87] DelayedMatrixStats_1.16.0 vctrs_0.3.8
[89] ellipsis_0.3.2 generics_0.1.1
[91] metap_1.7 tools_4.1.0
[93] beeswarm_0.4.0 munsell_0.5.0
[95] tweenr_1.0.2 DelayedArray_0.20.0
[97] fastmap_1.1.0 compiler_4.1.0
[99] abind_1.4-5 httpuv_1.6.5
[101] sessioninfo_1.2.2 plotly_4.10.0
[103] GenomeInfoDbData_1.2.7 gridExtra_2.3
[105] lattice_0.20-45 deldir_1.0-6
[107] mutoss_0.1-12 utf8_1.2.2
[109] later_1.3.0 jsonlite_1.7.2
[111] scales_1.1.1 ScaledMatrix_1.2.0
[113] pbapply_1.5-0 sparseMatrixStats_1.6.0
[115] renv_0.15.0-14 lazyeval_0.2.2
[117] promises_1.2.0.1 doParallel_1.0.16
[119] R.utils_2.11.0 goftest_1.2-3
[121] checkmate_2.0.0 spatstat.utils_2.3-0
[123] reticulate_1.22 sandwich_3.0-1
[125] rmarkdown_2.11 cowplot_1.1.1
[127] statmod_1.4.36 Rtsne_0.15
[129] EGSEAdata_1.22.0 uwot_0.1.11
[131] igraph_1.2.11 HDF5Array_1.22.1
[133] plotrix_3.8-2 numDeriv_2016.8-1.1
[135] survival_3.2-13 yaml_2.2.1
[137] htmltools_0.5.2 memoise_2.0.1
[139] locfit_1.5-9.4 graphlayouts_0.8.0
[141] viridisLite_0.4.0 digest_0.6.29
[143] assertthat_0.2.1 mime_0.12
[145] RSQLite_2.2.9 future.apply_1.8.1
[147] data.table_1.14.2 blob_1.2.2
[149] R.oo_1.24.0 labeling_0.4.2
[151] splines_4.1.0 rematch2_2.1.2
[153] Rhdf5lib_1.16.0 RCurl_1.98-1.6
[155] broom_0.7.11 hms_1.1.1
[157] modelr_0.1.8 rhdf5_2.38.0
[159] colorspace_2.0-2 mnormt_2.0.2
[161] BiocManager_1.30.16 tmvnsim_1.0-2
[163] ggbeeswarm_0.6.0 shape_1.4.6
[165] sass_0.4.0 Rcpp_1.0.7
[167] bookdown_0.24 RANN_2.6.1
[169] mvtnorm_1.1-3 circlize_0.4.13
[171] fansi_1.0.0 tzdb_0.2.0
[173] parallelly_1.30.0 R6_2.5.1
[175] grid_4.1.0 ggridges_0.5.3
[177] lifecycle_1.0.1 TFisher_0.2.0
[179] bluster_1.4.0 leiden_0.3.9
[181] jquerylib_0.1.4 safe_3.34.0
[183] Matrix_1.4-0 TH.data_1.1-0
[185] RcppAnnoy_0.0.19 RColorBrewer_1.1-2
[187] iterators_1.0.13 htmlwidgets_1.5.4
[189] beachmat_2.10.0 polyclip_1.10-0
[191] rvest_1.0.2 ComplexHeatmap_2.10.0
[193] mgcv_1.8-38 globals_0.14.0
[195] hgu133plus2.db_3.13.0 KEGGgraph_1.54.0
[197] codetools_0.2-18 lubridate_1.8.0
[199] metapod_1.2.0 gtools_3.9.2
[201] getPass_0.2-2 dbplyr_2.1.1
[203] RSpectra_0.16-0 R.methodsS3_1.8.1
[205] gtable_0.3.0 DBI_1.1.2
[207] git2r_0.29.0 highr_0.9
[209] tensor_1.5 httr_1.4.2
[211] KernSmooth_2.23-20 vroom_1.5.7
[213] stringi_1.7.6 reshape2_1.4.4
[215] farver_2.1.0 annotate_1.72.0
[217] viridis_0.6.2 Rgraphviz_2.38.0
[219] DT_0.20 xml2_1.3.3
[221] BiocNeighbors_1.12.0 scattermore_0.7
[223] bit_4.0.4 spatstat.data_2.1-2
[225] hgu133a.db_3.13.0 pkgconfig_2.0.3
[227] HTMLUtils_0.1.7 knitr_1.37