Analysis of scRNA-seq & CITE-seq Data Combined
Sub-cluster Macrophages (Round 2)
Jovana Maksimovic
December 19, 2022
Last updated: 2022-12-19
Checks: 7 0
Knit directory:
paed-cf-cite-seq/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20210524) was run prior to running the code in the R Markdown file.
Setting a seed ensures that any results that rely on randomness, e.g.
subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version e799f52. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for the
analysis have been committed to Git prior to generating the results (you can
use wflow_publish or wflow_git_commit). workflowr only
checks the R Markdown file, but you know if there are other scripts or data
files that it depends on. Below is the status of the Git repository when the
results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/obsolete/
Ignored: code/obsolete/
Ignored: data/190930_A00152_0150_BHTYCMDSXX/
Ignored: data/CellRanger/
Ignored: data/GSE127465_RAW/
Ignored: data/Homo_sapiens.gene_info
Ignored: data/SCEs/02_ZILIONIS.sct_normalised.SEU.rds
Ignored: data/SCEs/03_C133_Neeland.demultiplexed.SCE.rds
Ignored: data/SCEs/03_C133_Neeland.emptyDrops.SCE.rds
Ignored: data/SCEs/03_C133_Neeland.preprocessed.SCE.rds
Ignored: data/SCEs/03_CF_BAL_Pilot.CellRanger_v6.SCE.rds
Ignored: data/SCEs/03_CF_BAL_Pilot.emptyDrops.SCE.rds
Ignored: data/SCEs/03_CF_BAL_Pilot.preprocessed.SCE.rds
Ignored: data/SCEs/03_COMBO.clustered.SEU.rds
Ignored: data/SCEs/03_COMBO.clustered_annotated_macrophages_diet.SEU.rds
Ignored: data/SCEs/03_COMBO.clustered_annotated_others_diet.SEU.rds
Ignored: data/SCEs/03_COMBO.clustered_annotated_tcells_diet.SEU.rds
Ignored: data/SCEs/03_COMBO.clustered_diet.SEU.rds
Ignored: data/SCEs/03_COMBO.integrated.SEU.rds
Ignored: data/SCEs/03_COMBO.zilionis_mapped.SEU.rds
Ignored: data/SCEs/04_C133_Neeland.adt_dsb_normalised.rds
Ignored: data/SCEs/04_C133_Neeland.adt_integrated.rds
Ignored: data/SCEs/04_C133_Neeland.all_integrated.SEU.rds
Ignored: data/SCEs/04_CF_BAL_Pilot.CellRanger_v6.SCE.rds
Ignored: data/SCEs/04_CF_BAL_Pilot.emptyDrops.SCE.rds
Ignored: data/SCEs/04_CF_BAL_Pilot.preprocessed.SCE.rds
Ignored: data/SCEs/04_CF_BAL_Pilot.transfer_adt.SEU.rds
Ignored: data/SCEs/04_COMBO.clean_clustered.SEU.rds
Ignored: data/SCEs/04_COMBO.clean_clustered.SEU_bk.rds
Ignored: data/SCEs/04_COMBO.clean_integrated.SEU.rds
Ignored: data/SCEs/04_COMBO.clean_integrated.SEU_bk.rds
Ignored: data/SCEs/04_COMBO.clean_macrophages_diet.SEU.rds
Ignored: data/SCEs/04_COMBO.clean_others_diet.SEU.rds
Ignored: data/SCEs/04_COMBO.clean_tcells_diet.SEU.rds
Ignored: data/SCEs/04_COMBO.clustered.SEU.rds
Ignored: data/SCEs/04_COMBO.clustered_annotated_adt_diet.SEU.rds
Ignored: data/SCEs/04_COMBO.clustered_annotated_lung_diet.SEU.rds
Ignored: data/SCEs/04_COMBO.clustered_annotated_macrophages_diet.SEU.rds
Ignored: data/SCEs/04_COMBO.clustered_annotated_others_diet.SEU.rds
Ignored: data/SCEs/04_COMBO.clustered_annotated_tcells_diet.SEU.rds
Ignored: data/SCEs/04_COMBO.clustered_diet.SEU.rds
Ignored: data/SCEs/04_COMBO.integrated.SEU.rds
Ignored: data/SCEs/04_COMBO.macrophages_clustered.SEU.rds
Ignored: data/SCEs/04_COMBO.macrophages_integrated.SEU.rds
Ignored: data/SCEs/04_COMBO.others_clustered.SEU.rds
Ignored: data/SCEs/04_COMBO.others_integrated.SEU.rds
Ignored: data/SCEs/04_COMBO.tcells_clustered.SEU.rds
Ignored: data/SCEs/04_COMBO.tcells_integrated.SEU.rds
Ignored: data/SCEs/04_COMBO.zilionis_mapped.SEU.rds
Ignored: data/SCEs/05_CF_BAL_Pilot.transfer_adt.SEU.rds
Ignored: data/SCEs/05_COMBO.clean_clustered.SEU.rds
Ignored: data/SCEs/05_COMBO.clean_integrated.SEU.rds
Ignored: data/SCEs/05_COMBO.clean_macrophages_diet.SEU.rds
Ignored: data/SCEs/05_COMBO.clean_others_diet.SEU.rds
Ignored: data/SCEs/05_COMBO.clean_tcells_diet.SEU.rds
Ignored: data/SCEs/05_COMBO.clustered_annotated_adt_diet.SEU.rds
Ignored: data/SCEs/05_COMBO.clustered_annotated_lung_diet.SEU.rds
Ignored: data/SCEs/05_COMBO.clustered_annotated_macrophages_diet.SEU.rds
Ignored: data/SCEs/05_COMBO.clustered_annotated_others_diet.SEU.rds
Ignored: data/SCEs/05_COMBO.clustered_annotated_tcells_diet.SEU.rds
Ignored: data/SCEs/05_COMBO.macrophages_clustered.SEU.rds
Ignored: data/SCEs/05_COMBO.macrophages_integrated.SEU.rds
Ignored: data/SCEs/05_COMBO.others_clustered.SEU.rds
Ignored: data/SCEs/05_COMBO.others_integrated.SEU.rds
Ignored: data/SCEs/05_COMBO.tcells_clustered.SEU.rds
Ignored: data/SCEs/05_COMBO.tcells_integrated.SEU.rds
Ignored: data/SCEs/06_COMBO.clean_clustered.DIET.rds
Ignored: data/SCEs/06_COMBO.clean_clustered.SEU.rds
Ignored: data/SCEs/06_COMBO.clean_integrated.SEU.rds
Ignored: data/SCEs/06_COMBO.clean_macrophages_diet.SEU.rds
Ignored: data/SCEs/06_COMBO.clean_others_diet.SEU.rds
Ignored: data/SCEs/06_COMBO.clean_tcells_diet.SEU.rds
Ignored: data/SCEs/06_COMBO.macrophages_clustered.SEU.rds
Ignored: data/SCEs/06_COMBO.macrophages_clustered_dbl.SEU.rds
Ignored: data/SCEs/06_COMBO.macrophages_integrated.SEU.rds
Ignored: data/SCEs/06_COMBO.macrophages_integrated_dbl.SEU.rds
Ignored: data/SCEs/06_COMBO.others_clustered.SEU.rds
Ignored: data/SCEs/06_COMBO.others_integrated.SEU.rds
Ignored: data/SCEs/06_COMBO.tcells_clustered.SEU.rds
Ignored: data/SCEs/06_COMBO.tcells_integrated.SEU.rds
Ignored: data/SCEs/07_COMBO.macrophages_clustered.SEU.rds
Ignored: data/SCEs/07_COMBO.macrophages_integrated.SEU.rds
Ignored: data/SCEs/C133_Neeland.CellRanger.SCE.rds
Ignored: data/SCEs/experiment1_doublets.rds
Ignored: data/SCEs/experiment2_doublets.rds
Ignored: data/SCEs/obsolete/
Ignored: data/cellsnp-lite/
Ignored: data/emptyDrops/obsolete/
Ignored: data/obsolete/
Ignored: data/sample_sheets/obsolete/
Ignored: output/marker-analysis/obsolete/
Ignored: output/obsolete/
Ignored: rename_captures.R
Ignored: renv/library/
Ignored: renv/staging/
Ignored: wflow_background.R
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were made
to the R Markdown (analysis/08_COMBO.cluster_macrophages_round2.Rmd) and HTML (docs/08_COMBO.cluster_macrophages_round2.html)
files. If you’ve configured a remote Git repository (see
?wflow_git_remote), click on the hyperlinks in the table below to
view the files as they were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | e799f52 | Jovana Maksimovic | 2022-12-19 | wflow_publish(c("analysis/emptyDrops.Rmd", "analysis/postprocess*.Rmd", |
| html | 63f8ee8 | Jovana Maksimovic | 2022-12-15 | Build site. |
| Rmd | 916bafa | Jovana Maksimovic | 2022-12-15 | wflow_publish(c("analysis/.emptyDrops.Rmd", "analysis/postprocess_*.Rmd", |
| html | 4368d1d | Jovana Maksimovic | 2022-12-09 | Build site. |
| Rmd | 3e823ad | Jovana Maksimovic | 2022-12-09 | wflow_publish("analysis/08_COMBO.cluster_macrophages_round2.Rmd") |
1 Load libraries
2 Load Data
Load the clustered and labelled scRNA-seq and CITE-seq data.
seuInt <- readRDS(file = here("data/SCEs/06_COMBO.macrophages_clustered.SEU.rds"))
seuIntAn object of class Seurat
33301 features across 33161 samples within 3 assays
Active assay: integrated (3000 features, 3000 variable features)
2 other assays present: RNA, SCT
2 dimensional reductions calculated: pca, umap2.1 Load manual annotations
labels <- read_csv(here("data/macrophage_subcluster_annotation_29.05.22.csv"))
seuInt@meta.data %>%
left_join(labels %>%
mutate(Annotation = ifelse(is.na(Annotation),
"SUSPECT",
Annotation),
Broad = ifelse(is.na(Broad),
"SUSPECT",
Broad)) %>%
mutate(Cluster = as.factor(Cluster),
Annotation = as.factor(Annotation)),
by = c("integrated_snn_res.1" = "Cluster")) -> ann
ann %>% dplyr::pull(Annotation) -> seuInt$Annotation
ann %>% dplyr::pull(Broad) -> seuInt$Broad
seuInt$Annotation <- fct_drop(seuInt$Annotation)
seuInt$Broad <- fct_drop(seuInt$Broad)
seuIntAn object of class Seurat
33301 features across 33161 samples within 3 assays
Active assay: integrated (3000 features, 3000 variable features)
2 other assays present: RNA, SCT
2 dimensional reductions calculated: pca, umap2.2 Check for doublet enrichment
The macro-T cluster expresses both macrophage and T-cell marker genes so we need to check if it is artefactual e.g. contains doublets. We have already removed a total of 3826 heterogenic, cross-sample doublets based on vireo and hashedDrops calls. However, those methods cannot detect heterotypic and homotypic within-sample doublets. We have also run scds and scDblFinder to detect putative within-sample doublets.
Load doublet detection results and match up with annotated cells.
e1Doublets <- readRDS(here("data/SCEs/experiment1_doublets.rds"))
e1Doublets$cell <- paste0("A-", e1Doublets$cell)
e2Doublets <- readRDS(here("data/SCEs/experiment2_doublets.rds"))
e2Doublets$cell <- paste0("B-", e2Doublets$cell)
doublets <- rbind(e1Doublets, e2Doublets)
m <- match(colnames(seuInt), doublets$cell)
doublets <- doublets[m,]
all(doublets$cell == colnames(seuInt))[1] TRUEThe macro-T cluster is comprised of ~60% putative doublets.
table(doublets$scDblFinder.class == "doublet" & doublets$hybrid_call,
seuInt$Annotation) %>%
data.frame %>%
group_by(Var2) %>%
mutate(prop = Freq/sum(Freq)) %>%
ungroup() %>%
ggplot(aes(x = Var2, y = prop, fill = Var1)) +
geom_col() +
theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust = 1)) +
geom_hline(yintercept = 0.1, linetype = "dashed") +
labs(fill = "Doublet",
x = "Fine annotation",
y = "Proportion") -> p1
table(doublets$scDblFinder.class == "doublet" & doublets$hybrid_call,
seuInt$Annotation) %>%
data.frame %>%
ggplot(aes(x = Var2, y = Freq, fill = Var1)) +
geom_col() +
theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust = 1)) +
labs(fill = "Doublet",
x = "Fine annotation",
y = "Frequency") -> p2
(p2 | p1) + plot_layout(guides = "collect") &
theme(legend.position = "bottom")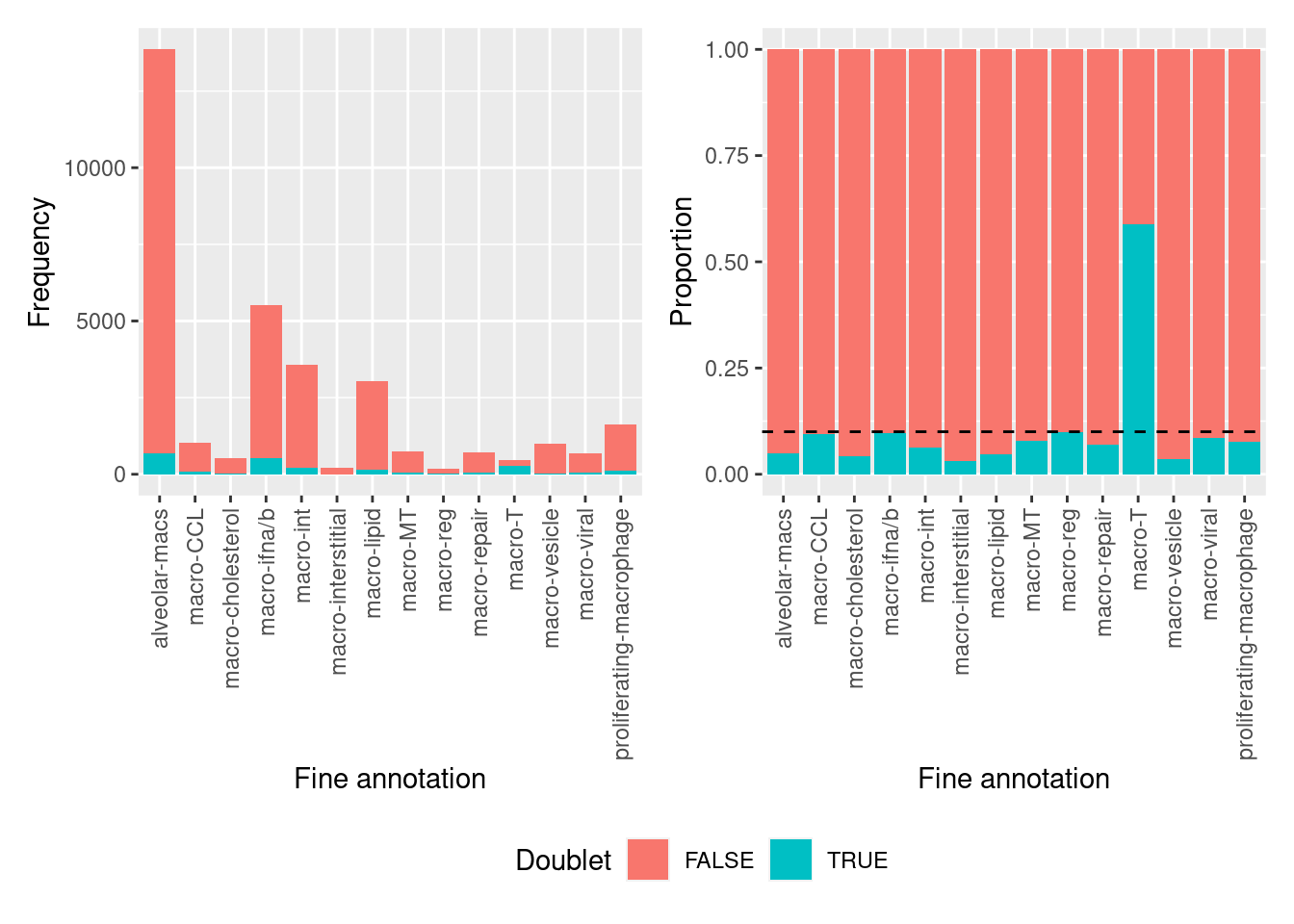
| Version | Author | Date |
|---|---|---|
| 4368d1d | Jovana Maksimovic | 2022-12-09 |
Calculate if doublets are statistically over-represented in any clusters using Fisher’s Exact Test.
tab <- table(doublets$scDblFinder.class == "doublet" & doublets$hybrid_call,
seuInt$Annotation)
dblStats <- table(doublets$scDblFinder.class == "doublet" & doublets$hybrid_call)
apply(tab, 2, function(x){
dblFreq <- matrix(c(x[2], dblStats[2] - x[2], x[1], dblStats[1] - x[1]),
nrow = 2,
dimnames = list(c("In cluster", "Not in cluster"),
c("Doublet", "Singlet")))
fisher.test(dblFreq, alternative = "greater")$p.value
}) -> pvals
pvals %>%
data.frame %>%
rownames_to_column(var = "cell") %>%
dplyr::rename("p.value" = ".") %>%
mutate(FDR = p.adjust(p.value, method = "BH")) %>%
ggplot(aes(y = -log10(FDR), x = cell,
fill = FDR < 0.05)) +
theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust = 1)) +
geom_col() 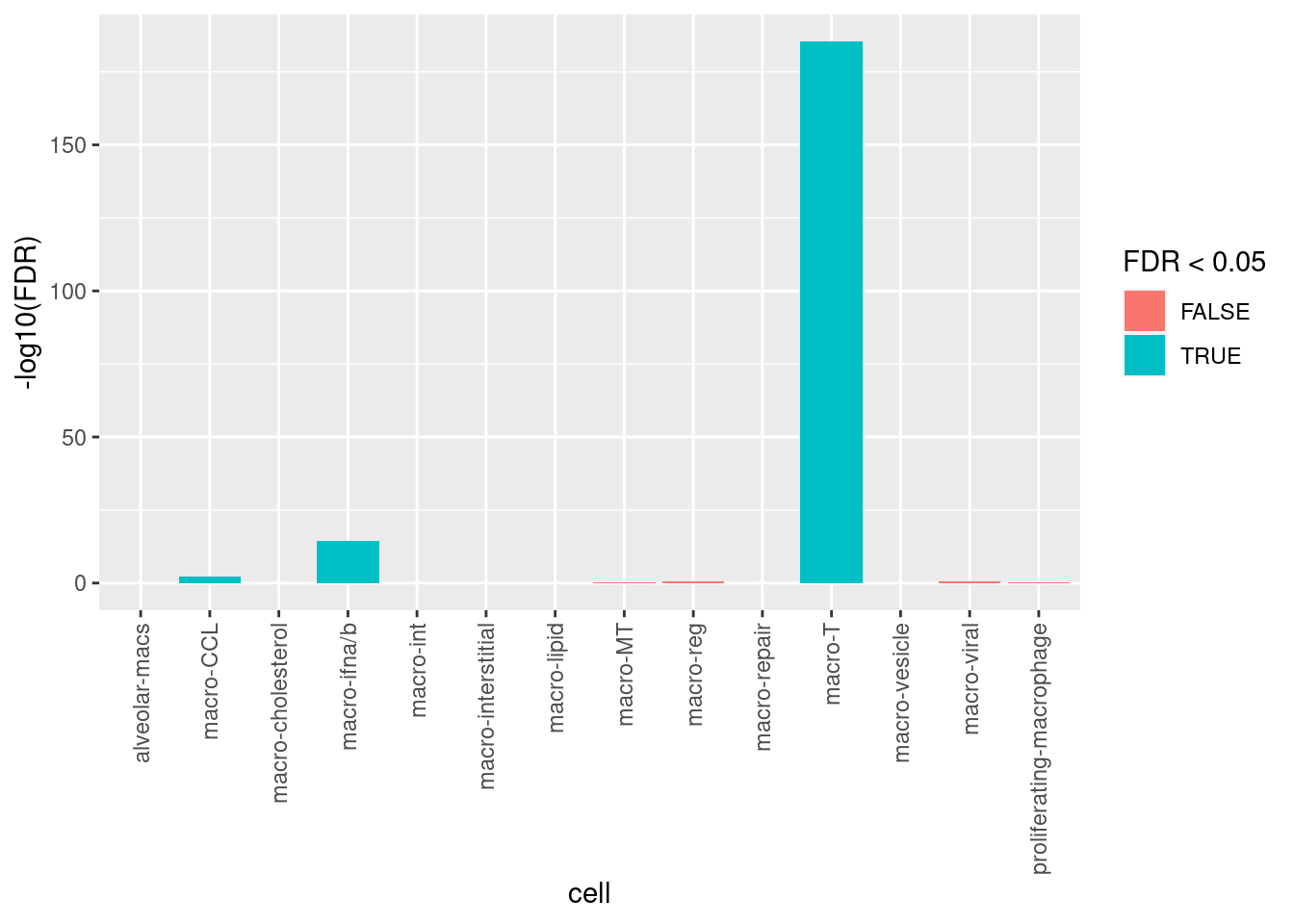
| Version | Author | Date |
|---|---|---|
| 4368d1d | Jovana Maksimovic | 2022-12-09 |
Doublets are defined as cells that are called doublets by both scds and scDblFinder. Filter out all doublets and all the cells in the macro-T cluster, which are all likely to be doublets based on association.
keep <- !(doublets$scDblFinder.class == "doublet" & doublets$hybrid_call)
DefaultAssay(seuInt) <- "RNA"
seu <- DietSeurat(subset(seuInt, cells = which(keep)),
assays = "RNA")
seuAn object of class Seurat
15578 features across 30847 samples within 1 assay
Active assay: RNA (15578 features, 0 variable features)rm(seuInt)
gc() used (Mb) gc trigger (Mb) max used (Mb)
Ncells 11665366 623.0 21569357 1152.0 18138030 968.7
Vcells 388405267 2963.3 2457345909 18748.1 2532459186 19321.23 Subcluster macrophages
Normalise and integrate data.
out <- here("data/SCEs/07_COMBO.macrophages_integrated.SEU.rds")
if(!file.exists(out)){
seuInt <- intDat(seu, type = "RNA",
reference = unique(seu$capture[seu$experiment == 1]))
saveRDS(seuInt, file = out)
} else {
seuInt <- readRDS(file = out)
}Visualise the data.
seuInt <- RunPCA(seuInt, verbose = FALSE, dims = 1:30) %>%
RunUMAP(verbose = FALSE, dims = 1:30)DimPlot(seuInt, group.by = "experiment", combine = FALSE)[[1]]
| Version | Author | Date |
|---|---|---|
| 4368d1d | Jovana Maksimovic | 2022-12-09 |
4 Clustering
4.1 Perform Linear Dimensional Reduction
p1 <- DimPlot(seuInt, reduction = "pca", group.by = "donor")
p2 <- DimPlot(seuInt, reduction = "pca", dims = c(1,3), group.by = "donor")
p3 <- DimPlot(seuInt, reduction = "pca", dims = c(2,3), group.by = "donor")
p4 <- DimPlot(seuInt, reduction = "pca", dims = c(3,4), group.by = "donor")
((p1 | p2) / (p3 | p4)) + plot_layout(guides = "collect") &
theme(legend.text = element_text(size = 8),
plot.title = element_text(size = 10),
axis.title = element_text(size = 9),
axis.text = element_text(size = 8))
| Version | Author | Date |
|---|---|---|
| 4368d1d | Jovana Maksimovic | 2022-12-09 |
DimHeatmap(seuInt, dims = 1:30, cells = 500, balanced = TRUE)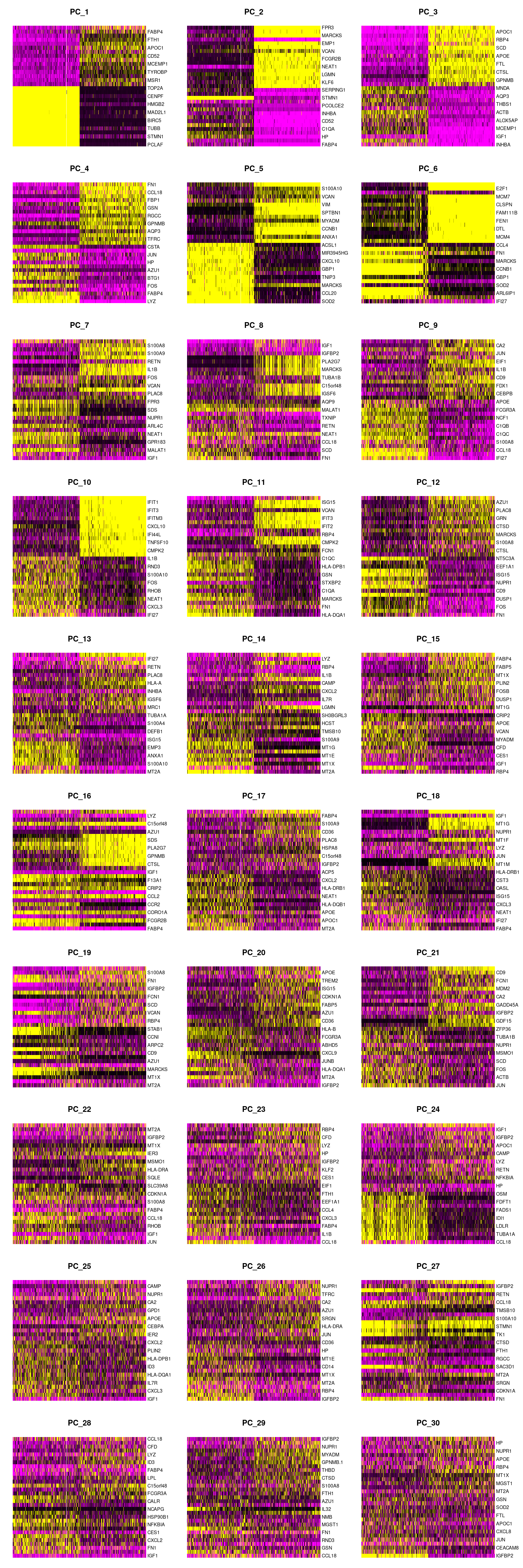
| Version | Author | Date |
|---|---|---|
| 4368d1d | Jovana Maksimovic | 2022-12-09 |
4.2 Determine the ‘Dimensionality’ of the Dataset
ElbowPlot(seuInt, ndims = 30)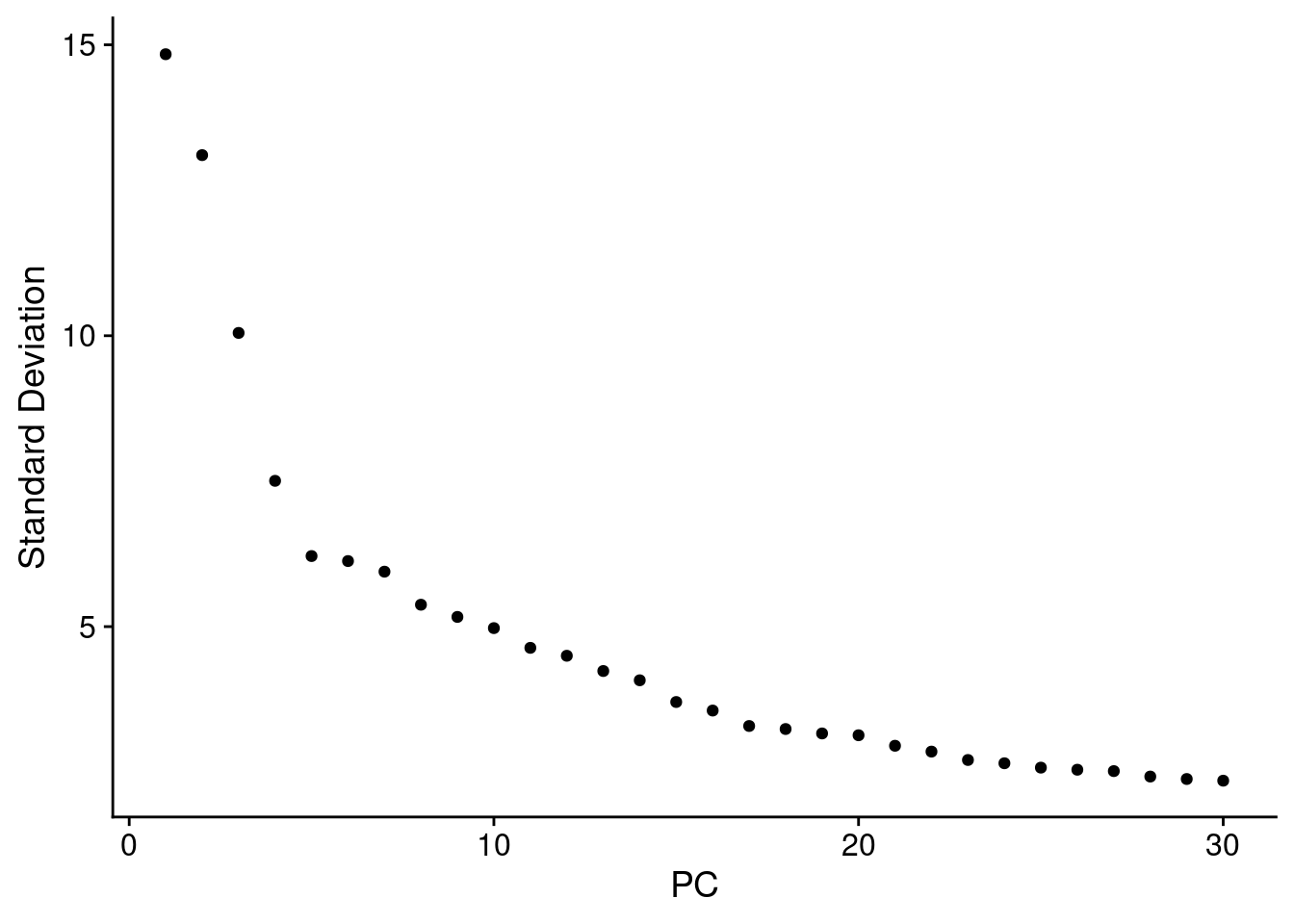
| Version | Author | Date |
|---|---|---|
| 4368d1d | Jovana Maksimovic | 2022-12-09 |
5 Cluster the Cells
Examine cluster number and size with respect to resolution.
out <- here("data/SCEs/07_COMBO.macrophages_clustered.SEU.rds")
if(!file.exists(out)){
seuInt <- FindNeighbors(seuInt, reduction = "pca", dims = 1:30)
seuInt <- FindClusters(seuInt, algorithm = 3,
resolution = seq(0.1, 1, by = 0.1))
seuInt <- RunUMAP(seuInt, dims = 1:10)
saveRDS(seuInt, file = out)
} else {
seuInt <- readRDS(file = out)
}
clustree::clustree(seuInt)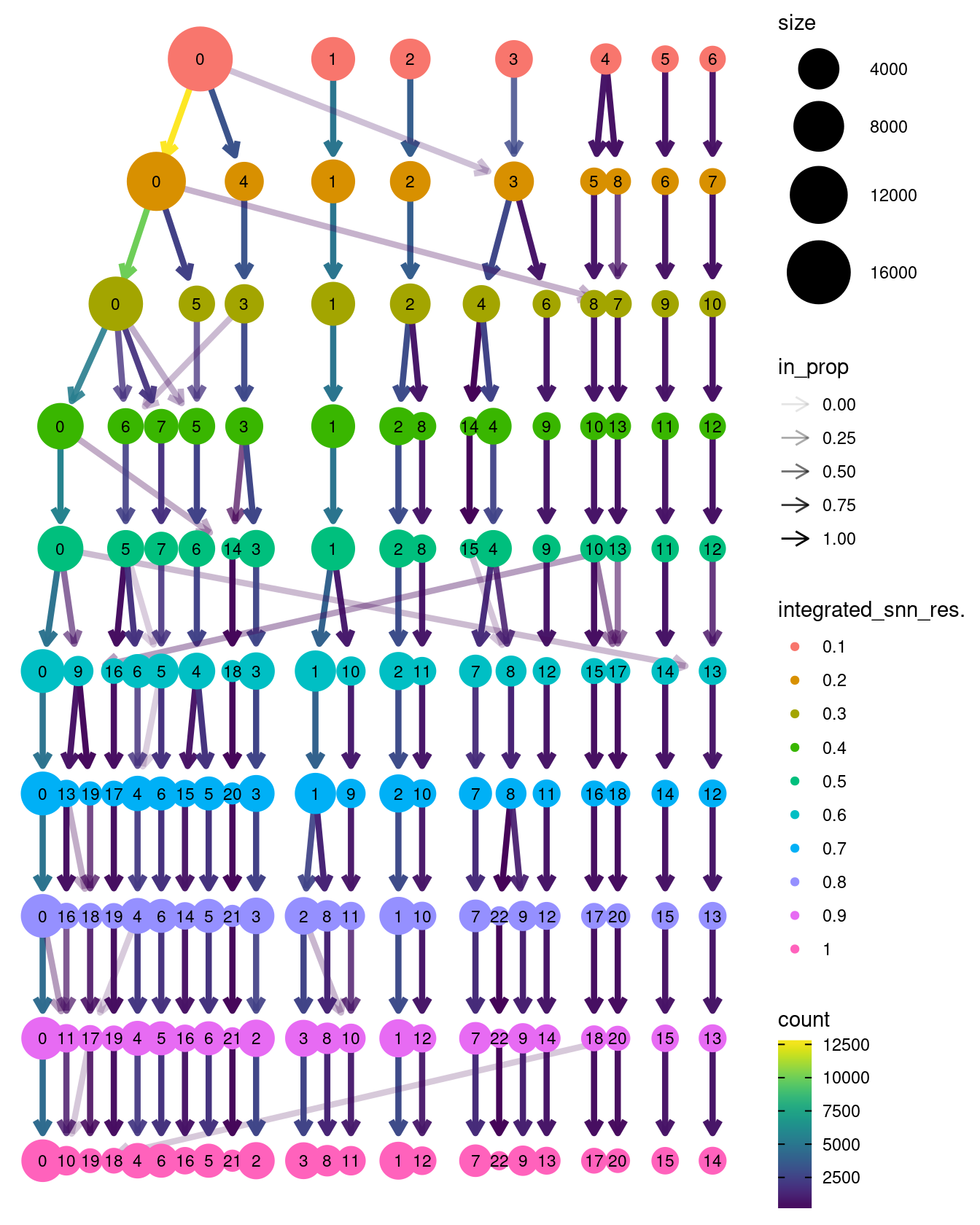
| Version | Author | Date |
|---|---|---|
| 4368d1d | Jovana Maksimovic | 2022-12-09 |
Choose a resolution. Visualise UMAP.
options(ggrepel.max.overlaps = Inf)
grp <- "integrated_snn_res.1"
DimPlot(seuInt, reduction = 'umap', label = TRUE, repel = FALSE,
label.size = 2.5, group.by = grp) + NoLegend()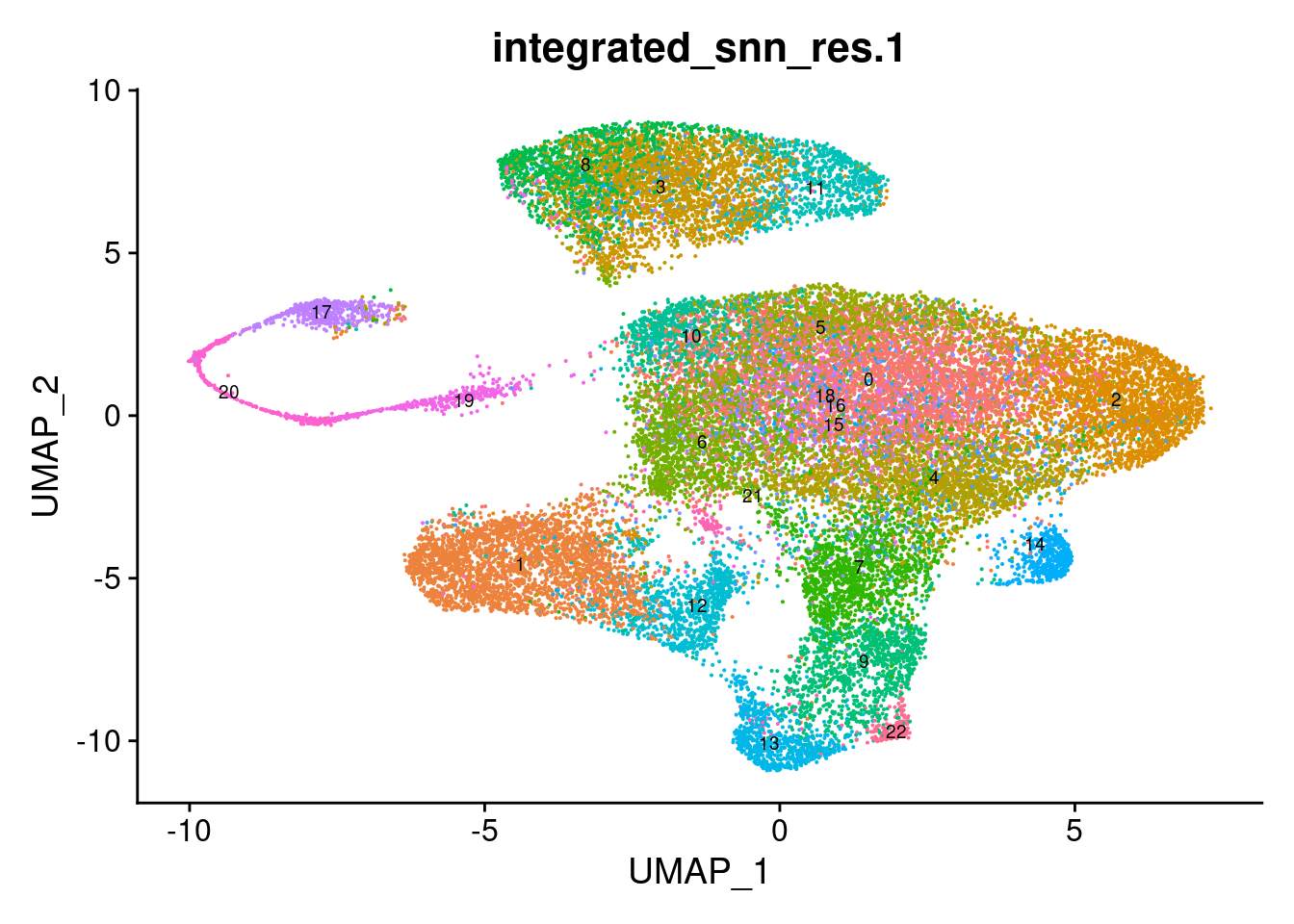
| Version | Author | Date |
|---|---|---|
| 4368d1d | Jovana Maksimovic | 2022-12-09 |
5.1 Examine clusters
Visualise quality metrics by cluster.
seuInt@meta.data %>%
ggplot(aes(x = integrated_snn_res.1,
y = predicted.annotation.l1.score,
fill = integrated_snn_res.1)) +
geom_violin(scale = "width") +
NoLegend() -> p1
seuInt@meta.data %>%
ggplot(aes(x = integrated_snn_res.1,
y = nCount_RNA,
fill = integrated_snn_res.1)) +
geom_violin(scale = "area") +
scale_y_log10() +
NoLegend() -> p2
seuInt@meta.data %>%
ggplot(aes(x = integrated_snn_res.1,
y = nFeature_RNA,
fill = integrated_snn_res.1)) +
geom_violin(scale = "area") +
scale_y_log10() +
NoLegend() -> p3
seuInt@meta.data %>%
ggplot(aes(x = integrated_snn_res.1,
y = predicted.ann_level_3.score,
fill = integrated_snn_res.1)) +
geom_violin(scale = "area") +
scale_y_log10() +
NoLegend() -> p4
((p1 | p2) / (p3 | p4)) & theme(text = element_text(size = 8))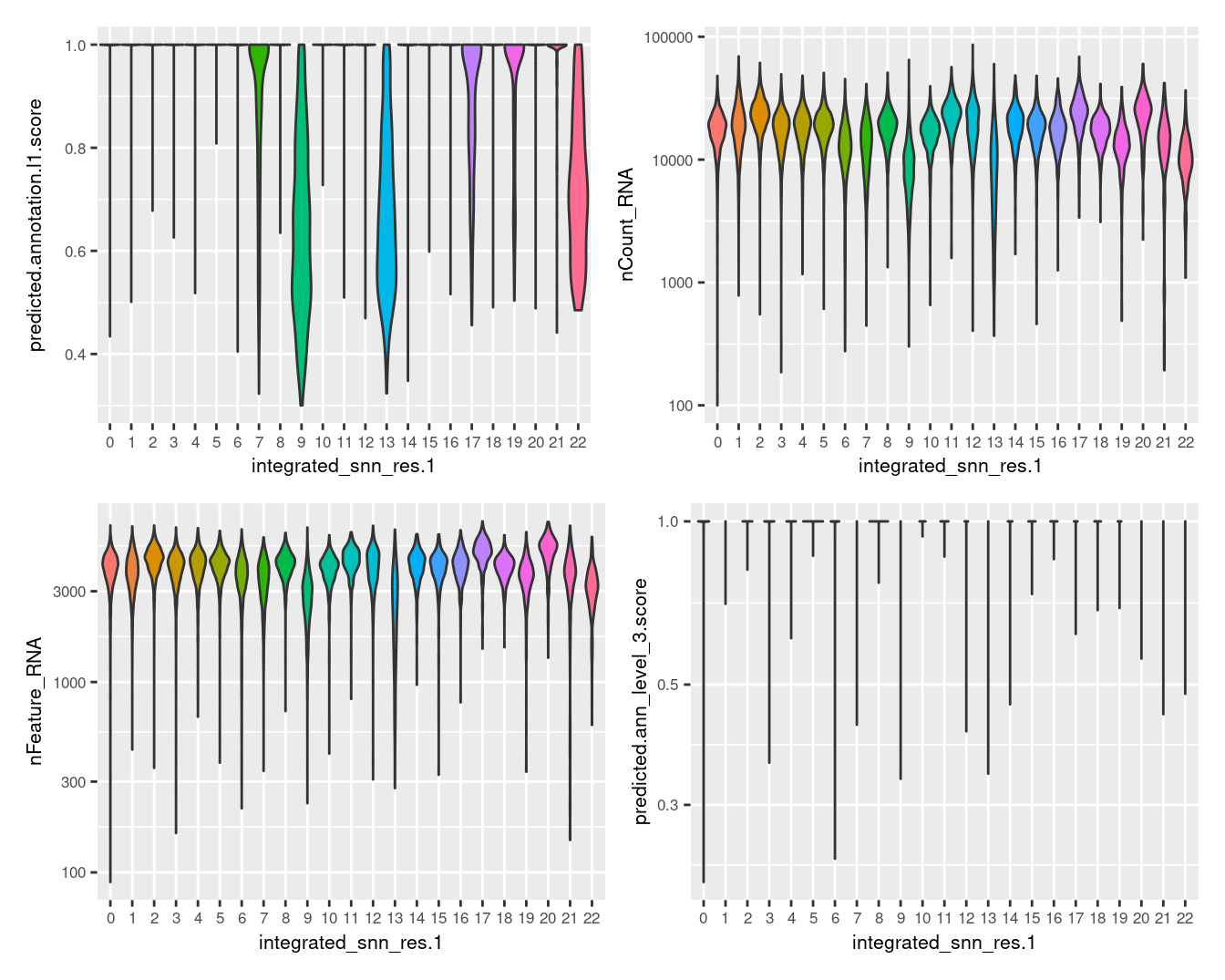
| Version | Author | Date |
|---|---|---|
| 4368d1d | Jovana Maksimovic | 2022-12-09 |
6 Identify Cluster Marker Genes
Adapted from Dr. Belinda Phipson’s work for (Sim et al. 2021).
6.1 Test for Marker Genes using limma
# limma-trend for DE
Idents(seuInt) <- grp
counts <- as.matrix(seuInt[["RNA"]]@counts)
y.org <- DGEList(counts)
logcounts <- normCounts(y.org, log = TRUE, prior.count = 0.5)
maxclust <- length(levels(Idents(seuInt))) - 1
clustgrp <- paste0("c", Idents(seuInt))
clustgrp <- factor(clustgrp, levels = paste0("c", 0:maxclust))
donor <- seuInt$donor
design <- model.matrix(~ 0 + clustgrp + donor)
colnames(design)[1:(length(levels(clustgrp)))] <- levels(clustgrp)
# Create contrast matrix
mycont <- matrix(NA, ncol = length(levels(clustgrp)),
nrow = length(levels(clustgrp)))
rownames(mycont) <- colnames(mycont) <- levels(clustgrp)
diag(mycont) <- 1
mycont[upper.tri(mycont)] <- -1/(length(levels(factor(clustgrp))) - 1)
mycont[lower.tri(mycont)] <- -1/(length(levels(factor(clustgrp))) - 1)
# Fill out remaining rows with 0s
zero.rows <- matrix(0, ncol = length(levels(clustgrp)),
nrow = (ncol(design) - length(levels(clustgrp))))
fullcont <- rbind(mycont, zero.rows)
rownames(fullcont) <- colnames(design)
fit <- lmFit(logcounts, design)
fit.cont <- contrasts.fit(fit, contrasts = fullcont)
fit.cont <- eBayes(fit.cont, trend = TRUE, robust = TRUE)
summary(decideTests(fit.cont)) c0 c1 c2 c3 c4 c5 c6 c7 c8 c9 c10 c11
Down 4674 6777 3587 2477 1989 2258 4080 4202 1798 6527 2478 2231
NotSig 8834 7185 8641 10873 11034 9848 7059 9681 10730 6938 11624 11405
Up 2070 1616 3350 2228 2555 3472 4439 1695 3050 2113 1476 1942
c12 c13 c14 c15 c16 c17 c18 c19 c20 c21 c22
Down 3484 6956 1317 1476 1297 758 1076 1921 1812 1890 2749
NotSig 10919 6439 12473 12964 11866 10163 12781 11362 8585 12120 10709
Up 1175 2183 1788 1138 2415 4657 1721 2295 5181 1568 21206.2 Test relative to a threshold (TREAT)
tr <- treat(fit.cont, fc = 1.5)
dt <- decideTests(tr)
summary(dt) c0 c1 c2 c3 c4 c5 c6 c7 c8 c9 c10 c11
Down 6 54 13 1 2 10 27 51 5 262 7 4
NotSig 15562 15483 15517 15566 15563 15526 15515 15501 15552 15211 15551 15543
Up 10 41 48 11 13 42 36 26 21 105 20 31
c12 c13 c14 c15 c16 c17 c18 c19 c20 c21 c22
Down 3 472 0 0 3 1 2 19 55 54 224
NotSig 15505 14924 15512 15569 15541 15375 15543 15518 14911 15520 15132
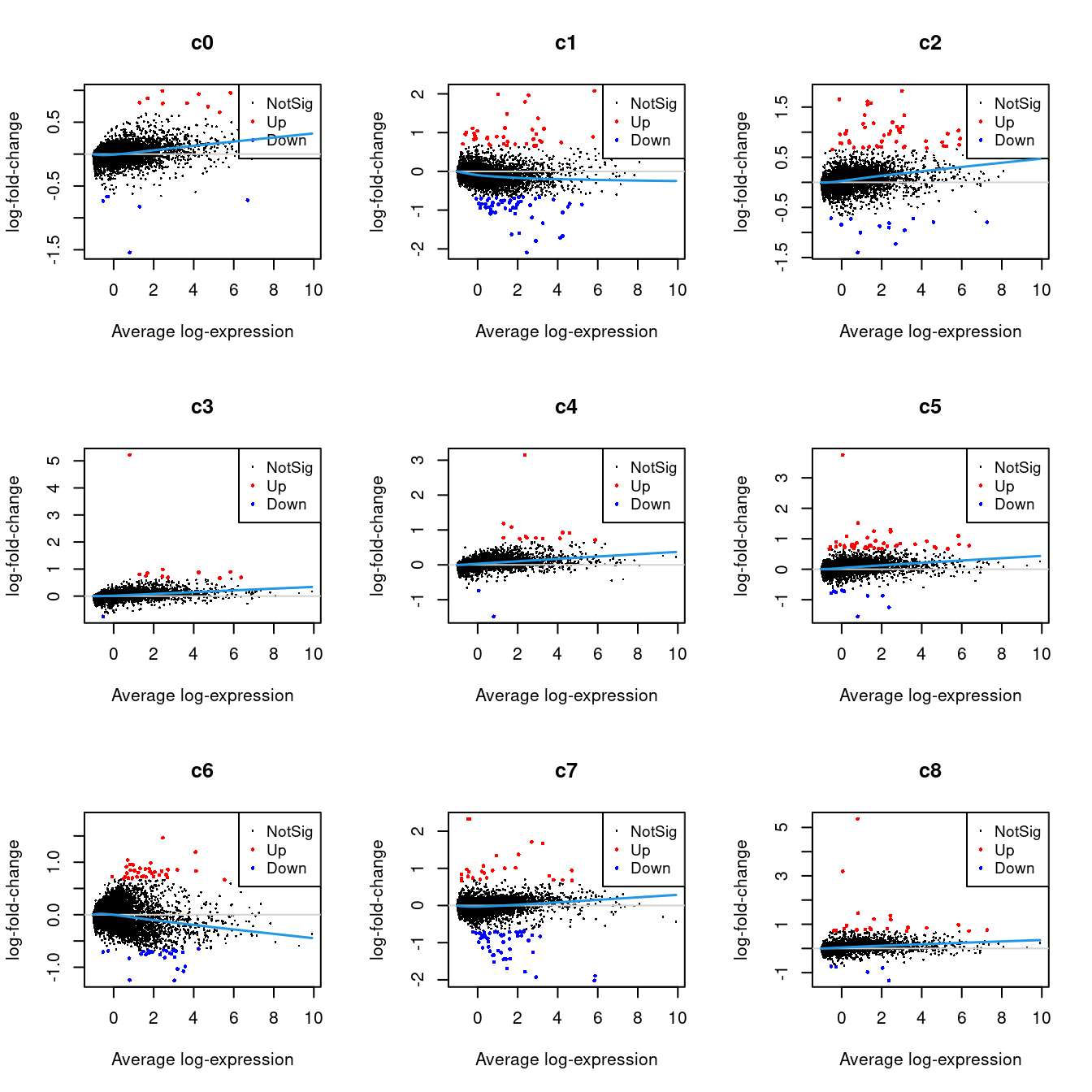
Up 70 182 66 9 34 202 33 41 612 4 2226.2.1 Mean-difference Plots per Cluster
par(mfrow=c(3,3))
for(i in 1:ncol(mycont)){
plotMD(tr, coef = i, status = dt[,i], hl.cex = 0.5)
abline(h = 0, col = "lightgrey")
lines(lowess(tr$Amean, tr$coefficients[,i]), lwd = 1.5, col = 4)
}
| Version | Author | Date |
|---|---|---|
| 4368d1d | Jovana Maksimovic | 2022-12-09 |
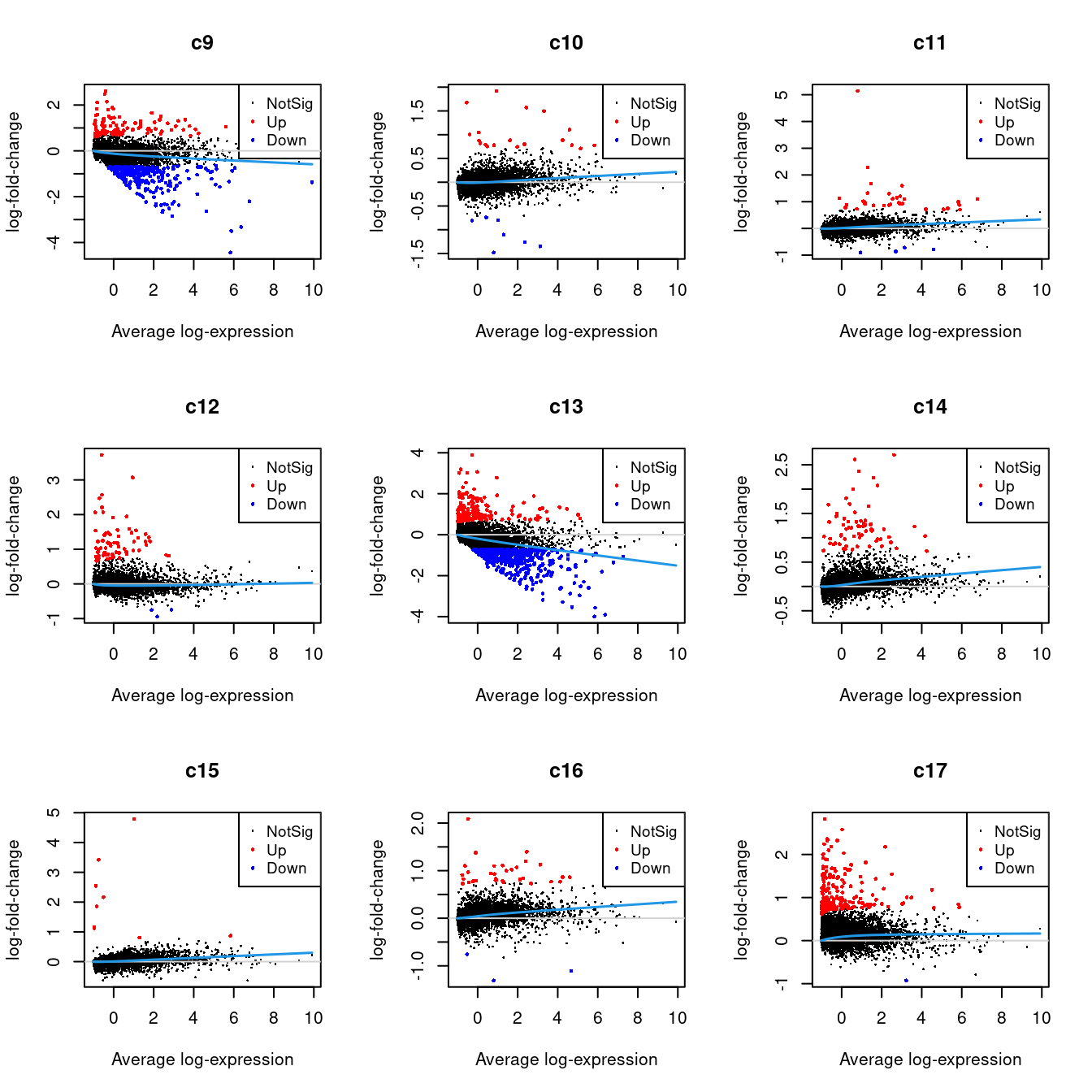
| Version | Author | Date |
|---|---|---|
| 4368d1d | Jovana Maksimovic | 2022-12-09 |
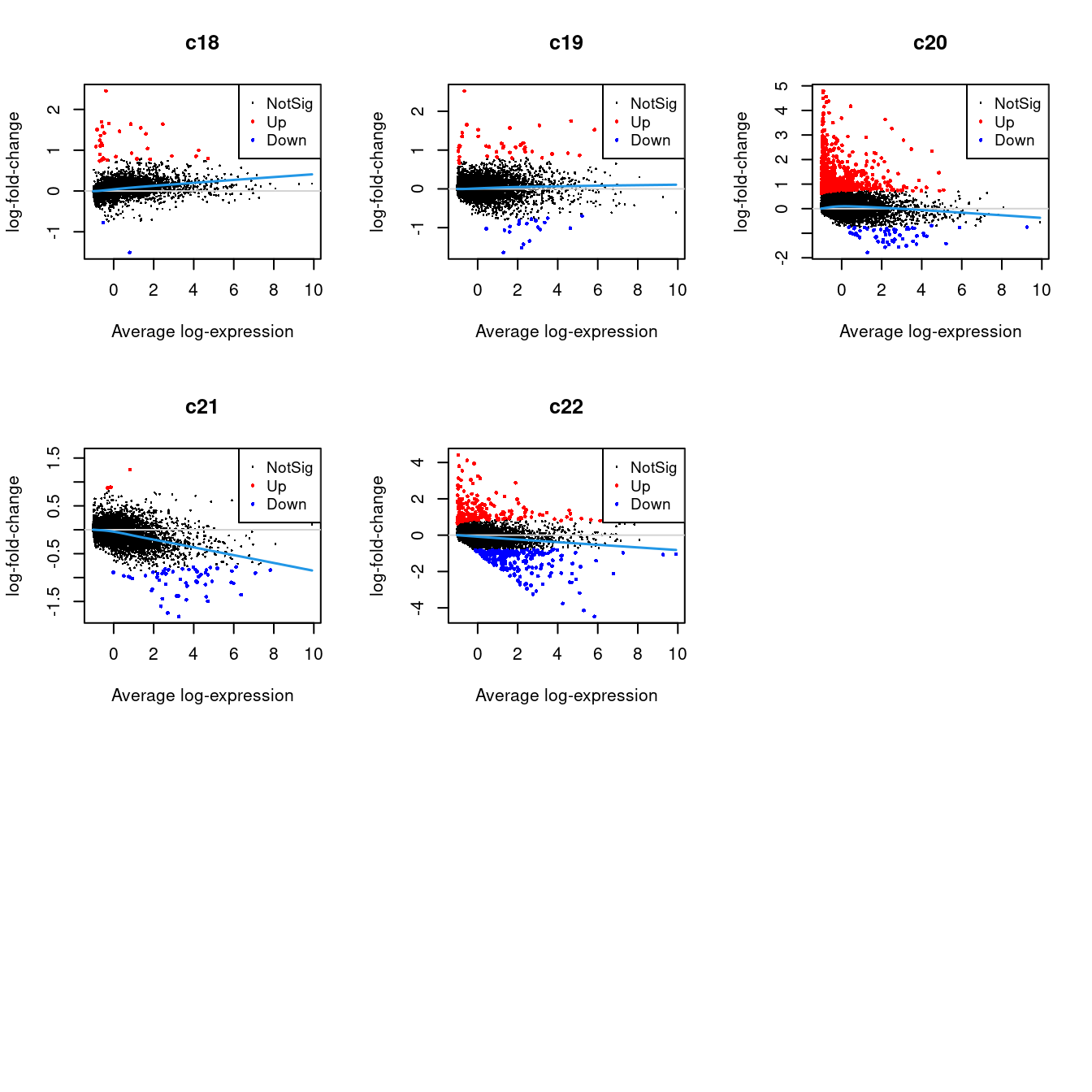
| Version | Author | Date |
|---|---|---|
| 4368d1d | Jovana Maksimovic | 2022-12-09 |
6.2.2 Export Marker Genes per cluster
options(scipen=-1, digits = 6)
contnames <- colnames(mycont)
dirName <- here("output/marker-analysis/06-COMBO-macrophages")
if(!dir.exists(dirName)) dir.create(dirName)
getCols <- setNames(c("SYMBOL","ENTREZID"),c("SYMBOL","ENTREZID"))
tr$genes <- data.frame(
lapply(getCols, function(column) {
mapIds(
x = org.Hs.eg.db,
keys = rownames(tr),
keytype = "SYMBOL",
column = column)
}),
row.names = rownames(tr))
gsAnnots <- buildIdx(entrezIDs = tr$genes$ENTREZID, species = "human",
msigdb.gsets = c("c2","c5"))[1] "Loading MSigDB Gene Sets ... "
[1] "Loaded gene sets for the collection c2 ..."
[1] "Indexed the collection c2 ..."
[1] "Created annotation for the collection c2 ..."
[1] "Loaded gene sets for the collection c5 ..."
[1] "Indexed the collection c5 ..."
[1] "Created annotation for the collection c5 ..."
[1] "Building KEGG pathways annotation object ... "reactomeIdx <-gsAnnots$c2@idx[grep("REACTOME",
names(gsAnnots$c2@idx))]
for(i in 1:length(contnames)){
top <- topTreat(tr, coef = i, n = Inf)
top <- top[top$logFC > 0, ]
write.csv(top[1:100, ],
file = glue("{dirName}/up-cluster-{contnames[i]}.csv"))
cameraPR(tr$t[,i], reactomeIdx) %>%
rownames_to_column(var = "Pathway") %>%
slice_head(n = 20) %>%
write_csv(file = here(glue("{dirName}/REACTOME-cluster-{contnames[i]}.csv")))
}6.2.3 Cluster marker gene dot plot
Genes duplicated between clusters are excluded.
sig.genes <- vector("list", ncol(tr))
p <- vector("list",length(sig.genes))
DefaultAssay(seuInt) <- "RNA"
for(i in 1:length(sig.genes)){
top <- topTreat(tr, coef = i, n = Inf)
sig.genes[[i]] <- rownames(top)[top$logFC > 0][1:10]
}
sig <- unlist(sig.genes)
geneCols <- c(rep(rep(c("grey","black"), each = 10), ncol(tr)/2),
rep("grey", 10))[!duplicated(sig)]
DotPlot(seuInt, features = sig[!duplicated(sig)],
group.by = "integrated_snn_res.1",
cols = c("lightgrey", "red"),
dot.scale = 3) +
RotatedAxis() +
FontSize(y.text = 8, x.text = 12) +
labs(y = element_blank(), x = element_blank()) +
coord_flip() +
theme(axis.text.y = element_text(color = geneCols)) +
ggtitle("Top 10 cluster marker genes without duplicates")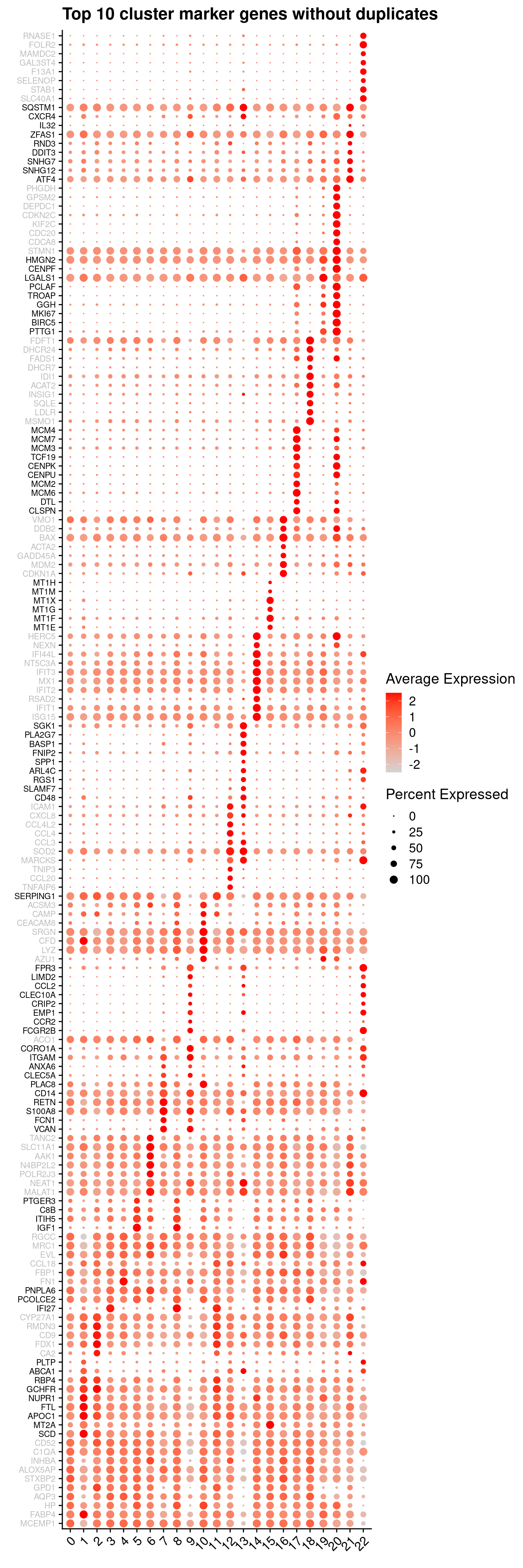
| Version | Author | Date |
|---|---|---|
| 4368d1d | Jovana Maksimovic | 2022-12-09 |
6.2.4 No. cells per cluster
seuInt@meta.data %>%
ggplot(aes(x = integrated_snn_res.1, fill = integrated_snn_res.1)) +
geom_bar() +
geom_text(aes(label = ..count..), stat = "count",
vjust = -0.5, colour = "black", size = 2) +
theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust = 1)) +
NoLegend()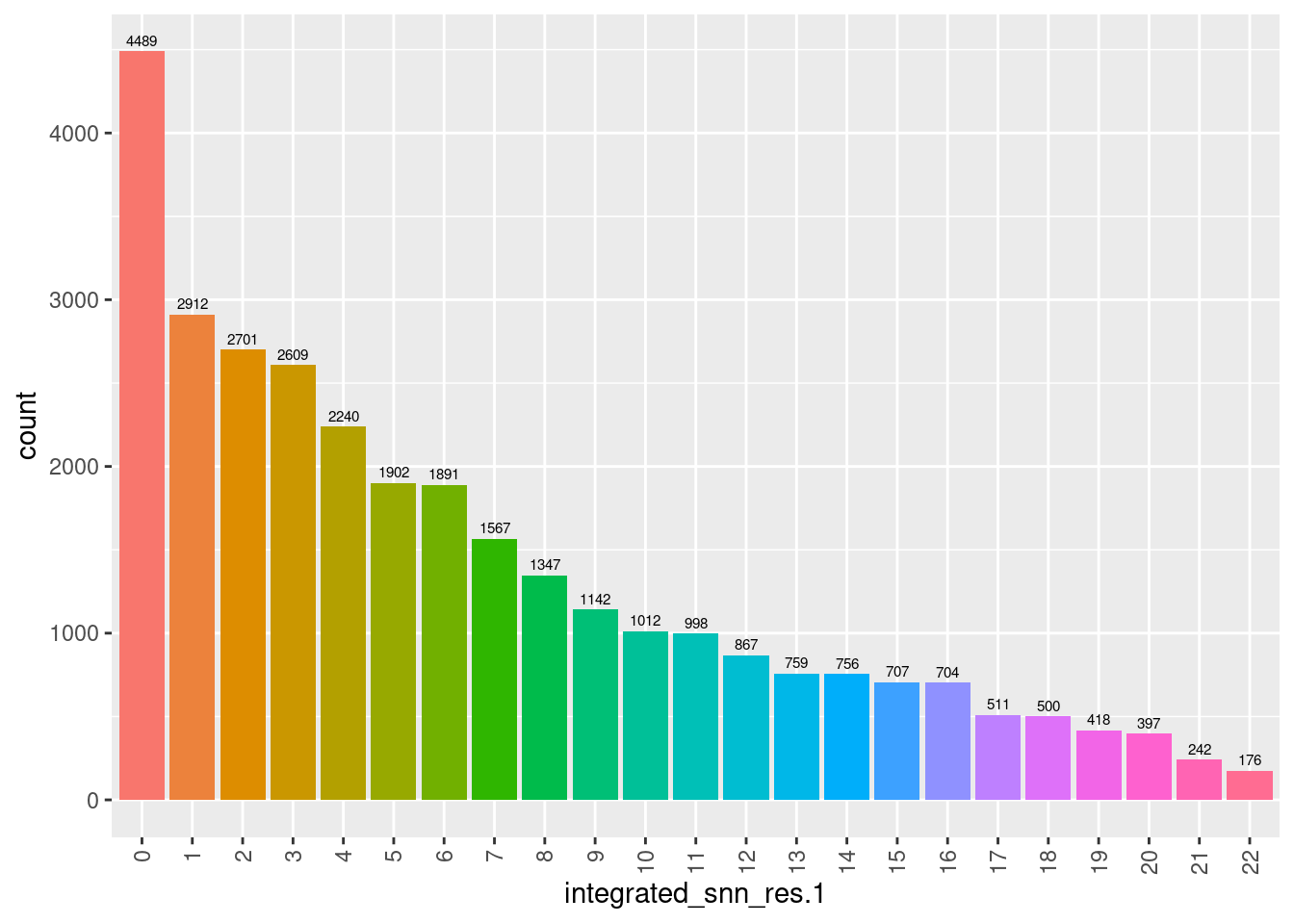
| Version | Author | Date |
|---|---|---|
| 4368d1d | Jovana Maksimovic | 2022-12-09 |
7 Load protein data
7.1 Add to Seurat object
seuAdt <- readRDS(here("data",
"SCEs",
"05_COMBO.clustered_annotated_adt_diet.SEU.rds"))
seuAdt <- subset(seuAdt, cells = colnames(seuInt))
all(colnames(seuAdt) == colnames(seuInt))[1] TRUEseuInt[["ADT.dsb"]] <- seuAdt[["ADT.dsb"]]
seuInt[["ADT.raw"]] <- seuAdt[["ADT.raw"]]
seuIntAn object of class Seurat
33440 features across 30847 samples within 5 assays
Active assay: RNA (15578 features, 0 variable features)
4 other assays present: SCT, integrated, ADT.dsb, ADT.raw
2 dimensional reductions calculated: pca, umaprm(seuAdt)
gc() used (Mb) gc trigger (Mb) max used (Mb)
Ncells 12136289 648.2 21569357 1152.0 21569357 1152.0
Vcells 2454265713 18724.6 5180913551 39527.3 5180905326 39527.27.2 Load protein annotations
prots <- read.csv(file = here("data",
"sample_sheets",
"TotalSeq-A_Universal_Cocktail_v1.0.csv")) %>%
dplyr::filter(grepl("^A0", id)) %>%
dplyr::filter(!grepl("[Ii]sotype", name)) 7.3 Visualise all ADTs
Normalised with DSB. CITE-seq ADT data was transferred to scRNA-seq using reference mapping and transfer.
cbind(seuInt@meta.data,
as.data.frame(t(seuInt@assays$ADT.dsb@data))) %>%
dplyr::group_by(integrated_snn_res.1, experiment) %>%
dplyr::summarize_at(.vars = prots$id, .funs = median) %>%
pivot_longer(c(-integrated_snn_res.1, -experiment), names_to = "ADT",
values_to = "ADT Exp.") %>%
left_join(prots, by = c("ADT" = "id")) %>%
mutate(Cluster = as.character(integrated_snn_res.1)) %>%
dplyr::rename(Protein = name) |>
dplyr::filter(experiment == 2) |>
ungroup() -> dat
plot(density(dat$`ADT Exp.`))
topMax <- 8
abline(v = topMax, lty = 2, col = "grey")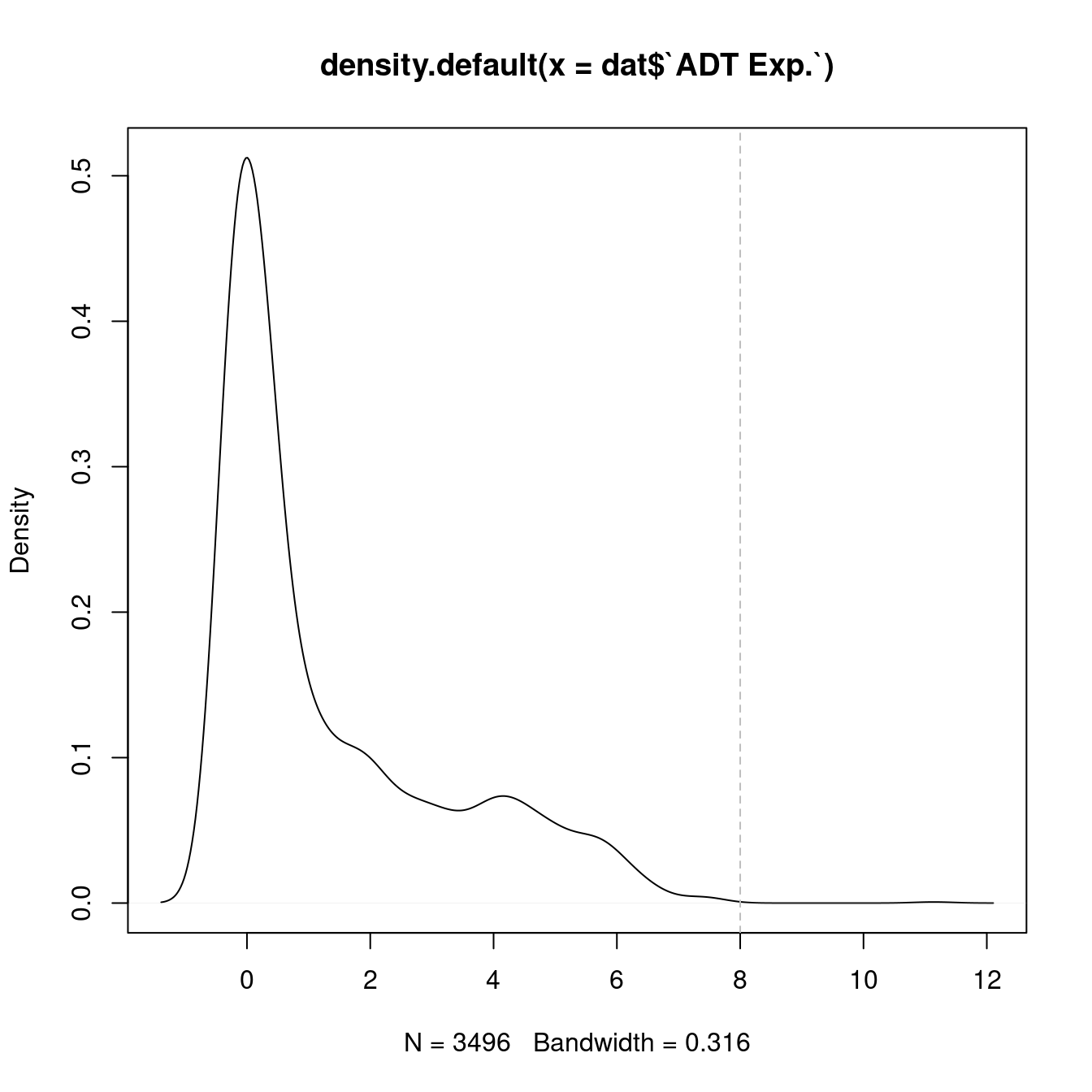
| Version | Author | Date |
|---|---|---|
| 4368d1d | Jovana Maksimovic | 2022-12-09 |
dat |>
heatmap(
.column = Cluster,
.row = Protein,
.value = `ADT Exp.`,
scale = "none",
rect_gp = grid::gpar(col = "white", lwd = 1),
show_row_names = TRUE,
column_names_gp = grid::gpar(fontsize = 10),
column_title_gp = grid::gpar(fontsize = 12),
row_names_gp = grid::gpar(fontsize = 8),
row_title_gp = grid::gpar(fontsize = 12),
column_title_side = "top",
palette_value = circlize::colorRamp2(seq(-1, topMax, length.out = 256),
viridis::magma(256)),
heatmap_legend_param = list(direction = "vertical")) 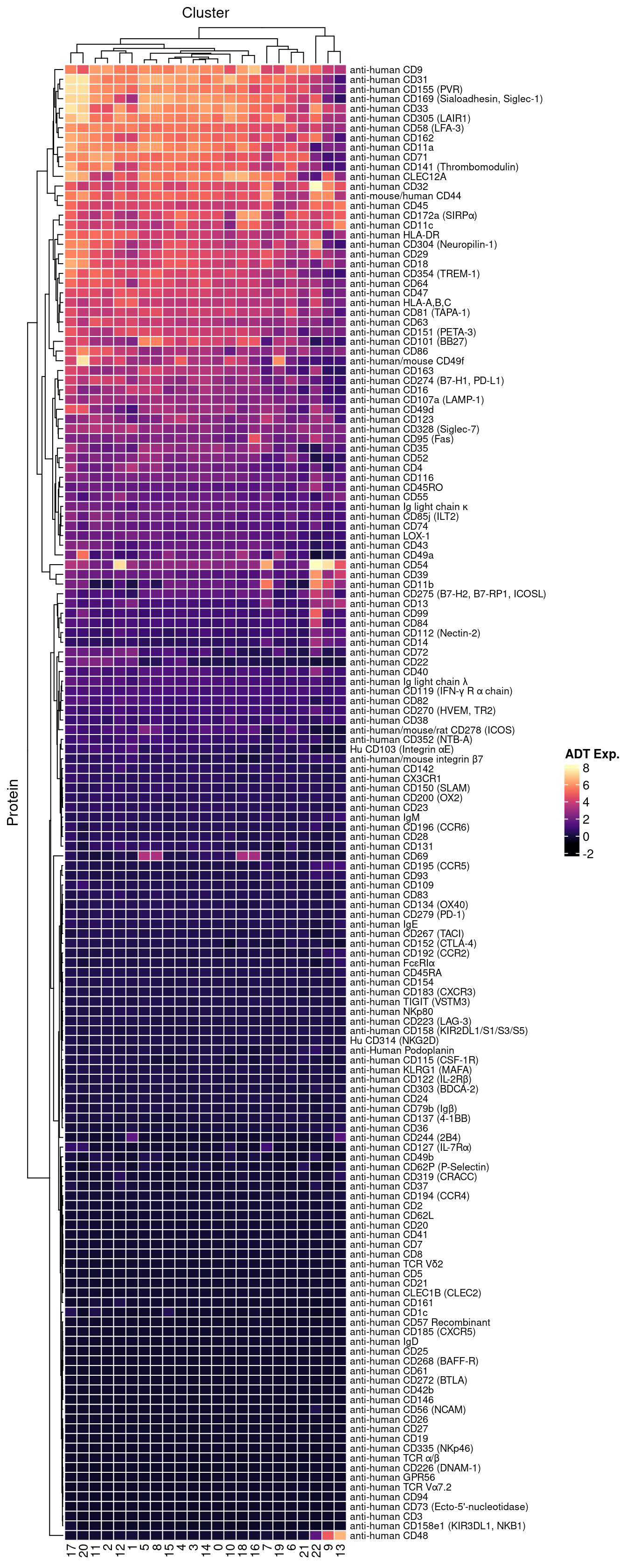
| Version | Author | Date |
|---|---|---|
| 4368d1d | Jovana Maksimovic | 2022-12-09 |
7.4 Visualise ADTs of interest
adt <- read_csv(file = here("data/Proteins_macs_22.04.22.csv"))
adt <- adt[!duplicated(adt$DNA_ID),]
dat %>%
inner_join(adt, by = c("ADT" = "DNA_ID")) %>%
dplyr::mutate(Protein = `Name for heatmap`) |>
heatmap(
.column = Cluster,
.row = Protein,
.value = `ADT Exp.`,
scale = "none",
rect_gp = grid::gpar(col = "white", lwd = 1),
show_row_names = TRUE,
column_names_gp = grid::gpar(fontsize = 10),
column_title_gp = grid::gpar(fontsize = 12),
row_names_gp = grid::gpar(fontsize = 8),
row_title_gp = grid::gpar(fontsize = 12),
column_title_side = "top",
palette_value = circlize::colorRamp2(seq(-1, topMax, length.out = 256),
viridis::magma(256)),
heatmap_legend_param = list(direction = "vertical")) 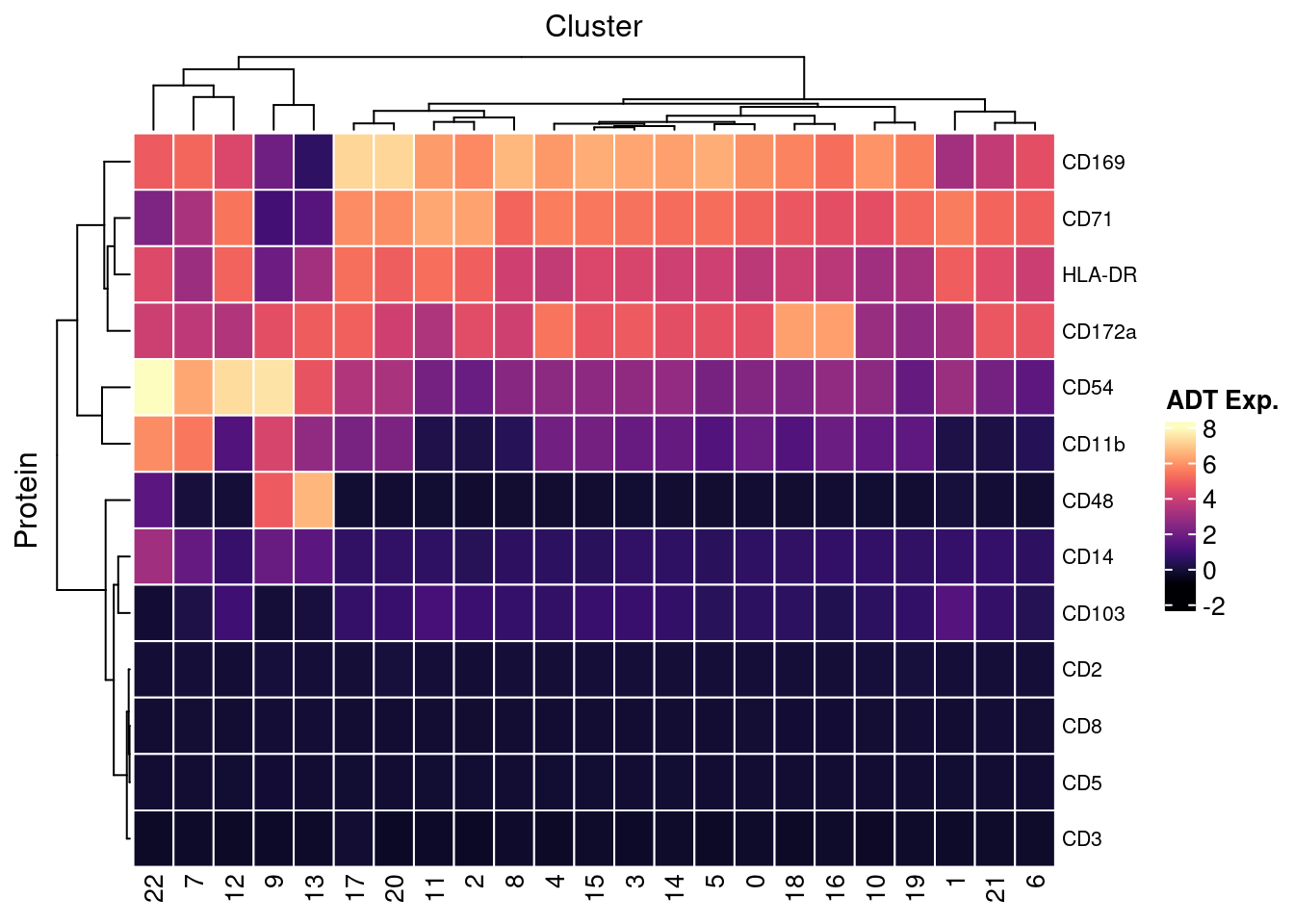
| Version | Author | Date |
|---|---|---|
| 4368d1d | Jovana Maksimovic | 2022-12-09 |
7.5 Visualise cytokines of interest
markers <- read_csv(file = here("data",
"macrophage_subcluster_cytokines.csv"),
col_names = FALSE)
p <- DotPlot(seuInt,
features = markers$X1,
cols = c("grey", "red"),
dot.scale = 5,
assay = "RNA",
group.by = "integrated_snn_res.1") +
theme(axis.text.x = element_text(angle = 90,
hjust = 1,
vjust = 0.5,
size = 8),
axis.text.y = element_text(size = 8),
text = element_text(size = 8)) +
coord_flip() +
labs(y = "Cluster", x = "Cytokine")
p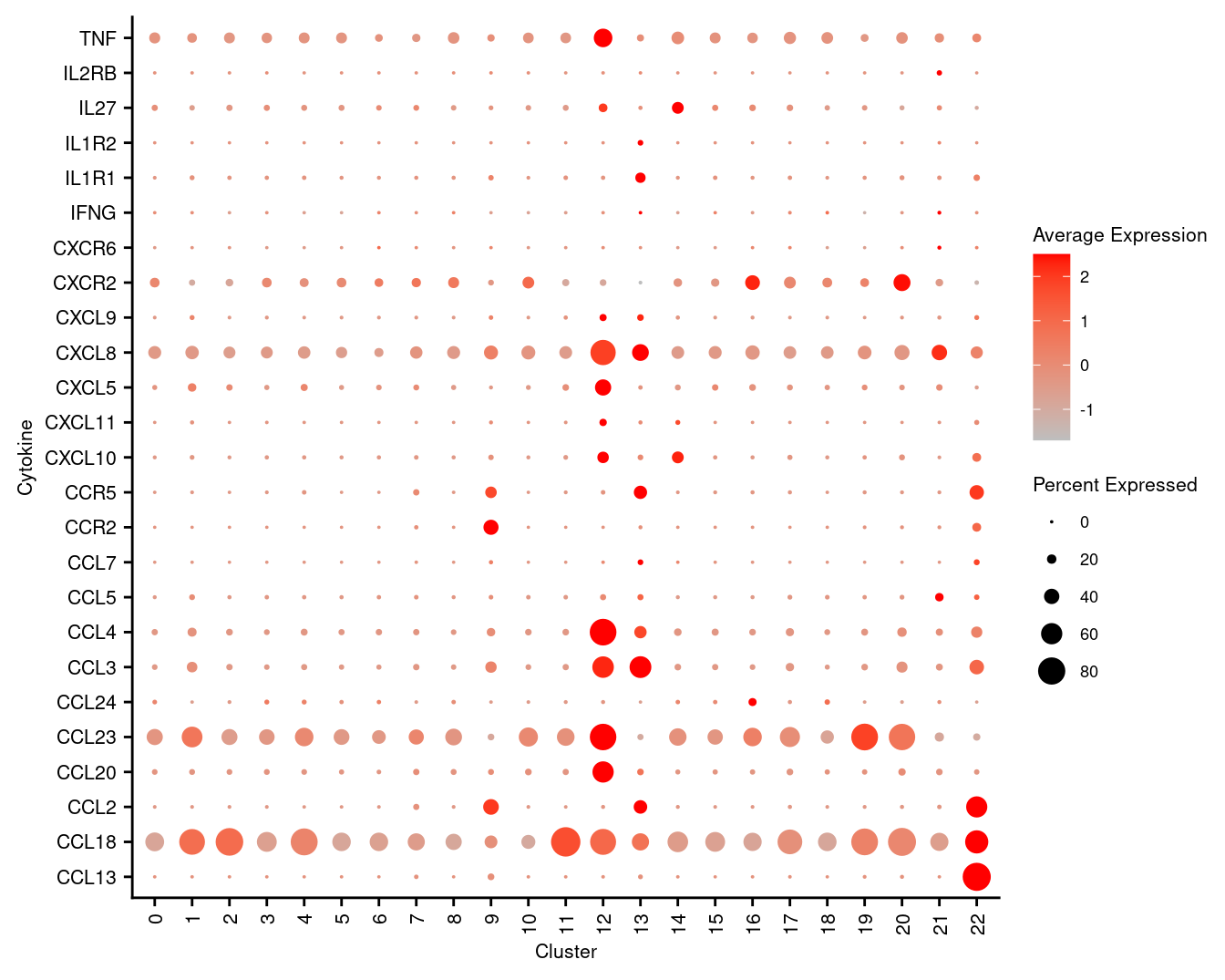
| Version | Author | Date |
|---|---|---|
| 4368d1d | Jovana Maksimovic | 2022-12-09 |
8 Session info
sessioninfo::session_info()─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.1.0 (2021-05-18)
os CentOS Linux 7 (Core)
system x86_64, linux-gnu
ui X11
language (EN)
collate en_AU.UTF-8
ctype en_AU.UTF-8
tz Australia/Melbourne
date 2022-12-19
pandoc 2.17.1.1 @ /usr/lib/rstudio-server/bin/quarto/bin/ (via rmarkdown)
─ Packages ───────────────────────────────────────────────────────────────────
! package * version date (UTC) lib source
P abind 1.4-5 2016-07-21 [?] CRAN (R 4.1.0)
P annotate 1.72.0 2021-10-26 [?] Bioconductor
P AnnotationDbi * 1.56.2 2021-11-09 [?] Bioconductor
P assertthat 0.2.1 2019-03-21 [?] CRAN (R 4.1.0)
P backports 1.4.1 2021-12-13 [?] CRAN (R 4.1.0)
P beachmat 2.10.0 2021-10-26 [?] Bioconductor
P beeswarm 0.4.0 2021-06-01 [?] CRAN (R 4.1.0)
P Biobase * 2.54.0 2021-10-26 [?] Bioconductor
P BiocGenerics * 0.40.0 2021-10-26 [?] Bioconductor
P BiocManager 1.30.16 2021-06-15 [?] CRAN (R 4.1.0)
P BiocNeighbors 1.12.0 2021-10-26 [?] Bioconductor
P BiocParallel * 1.28.3 2021-12-09 [?] Bioconductor
P BiocSingular 1.10.0 2021-10-26 [?] Bioconductor
P BiocStyle * 2.22.0 2021-10-26 [?] Bioconductor
P Biostrings 2.62.0 2021-10-26 [?] Bioconductor
P bit 4.0.4 2020-08-04 [?] CRAN (R 4.1.0)
P bit64 4.0.5 2020-08-30 [?] CRAN (R 4.0.2)
P bitops 1.0-7 2021-04-24 [?] CRAN (R 4.0.2)
P blob 1.2.2 2021-07-23 [?] CRAN (R 4.1.0)
P bluster 1.4.0 2021-10-26 [?] Bioconductor
P bookdown 0.24 2021-09-02 [?] CRAN (R 4.1.0)
P broom 0.7.11 2022-01-03 [?] CRAN (R 4.1.0)
P bslib 0.3.1 2021-10-06 [?] CRAN (R 4.1.0)
P cachem 1.0.6 2021-08-19 [?] CRAN (R 4.1.0)
P callr 3.7.0 2021-04-20 [?] CRAN (R 4.1.0)
P caTools 1.18.2 2021-03-28 [?] CRAN (R 4.1.0)
P cellranger 1.1.0 2016-07-27 [?] CRAN (R 4.1.0)
P checkmate 2.0.0 2020-02-06 [?] CRAN (R 4.0.2)
P circlize 0.4.13 2021-06-09 [?] CRAN (R 4.1.0)
P cli 3.1.0 2021-10-27 [?] CRAN (R 4.1.0)
P clue 0.3-60 2021-10-11 [?] CRAN (R 4.1.0)
P cluster 2.1.2 2021-04-17 [?] CRAN (R 4.1.0)
P clustree * 0.4.4 2021-11-08 [?] CRAN (R 4.1.0)
P codetools 0.2-18 2020-11-04 [?] CRAN (R 4.1.0)
P colorspace 2.0-2 2021-06-24 [?] CRAN (R 4.0.2)
P ComplexHeatmap 2.10.0 2021-10-26 [?] Bioconductor
P cowplot 1.1.1 2020-12-30 [?] CRAN (R 4.0.2)
P crayon 1.4.2 2021-10-29 [?] CRAN (R 4.1.0)
P data.table 1.14.2 2021-09-27 [?] CRAN (R 4.1.0)
P DBI 1.1.2 2021-12-20 [?] CRAN (R 4.1.0)
P dbplyr 2.1.1 2021-04-06 [?] CRAN (R 4.1.0)
P DelayedArray 0.20.0 2021-10-26 [?] Bioconductor
P DelayedMatrixStats 1.16.0 2021-10-26 [?] Bioconductor
P deldir 1.0-6 2021-10-23 [?] CRAN (R 4.1.0)
P dendextend 1.15.2 2021-10-28 [?] CRAN (R 4.1.0)
P digest 0.6.29 2021-12-01 [?] CRAN (R 4.1.0)
P doParallel 1.0.16 2020-10-16 [?] CRAN (R 4.0.2)
P doRNG 1.8.2 2020-01-27 [?] CRAN (R 4.1.0)
P dplyr * 1.0.7 2021-06-18 [?] CRAN (R 4.1.0)
P dqrng 0.3.0 2021-05-01 [?] CRAN (R 4.1.0)
P DropletUtils * 1.14.1 2021-11-08 [?] Bioconductor
P DT 0.20 2021-11-15 [?] CRAN (R 4.1.0)
P edgeR * 3.36.0 2021-10-26 [?] Bioconductor
P EGSEA * 1.22.0 2021-10-26 [?] Bioconductor
P EGSEAdata 1.22.0 2021-10-30 [?] Bioconductor
P ellipsis 0.3.2 2021-04-29 [?] CRAN (R 4.0.2)
P evaluate 0.14 2019-05-28 [?] CRAN (R 4.0.2)
P fansi 1.0.0 2022-01-10 [?] CRAN (R 4.1.0)
P farver 2.1.0 2021-02-28 [?] CRAN (R 4.0.2)
P fastmap 1.1.0 2021-01-25 [?] CRAN (R 4.1.0)
P fitdistrplus 1.1-6 2021-09-28 [?] CRAN (R 4.1.0)
P forcats * 0.5.1 2021-01-27 [?] CRAN (R 4.1.0)
P foreach 1.5.1 2020-10-15 [?] CRAN (R 4.0.2)
P fs 1.5.2 2021-12-08 [?] CRAN (R 4.1.0)
P future 1.23.0 2021-10-31 [?] CRAN (R 4.1.0)
P future.apply 1.8.1 2021-08-10 [?] CRAN (R 4.1.0)
P gage * 2.44.0 2021-10-26 [?] Bioconductor
P generics 0.1.1 2021-10-25 [?] CRAN (R 4.1.0)
GenomeInfoDb * 1.30.1 2022-01-30 [1] Bioconductor
P GenomeInfoDbData 1.2.7 2021-12-21 [?] Bioconductor
P GenomicRanges * 1.46.1 2021-11-18 [?] Bioconductor
P GetoptLong 1.0.5 2020-12-15 [?] CRAN (R 4.0.2)
P getPass 0.2-2 2017-07-21 [?] CRAN (R 4.0.2)
P ggbeeswarm 0.6.0 2017-08-07 [?] CRAN (R 4.1.0)
P ggforce 0.3.3 2021-03-05 [?] CRAN (R 4.1.0)
P ggplot2 * 3.3.5 2021-06-25 [?] CRAN (R 4.0.2)
P ggraph * 2.0.5 2021-02-23 [?] CRAN (R 4.1.0)
P ggrepel 0.9.1 2021-01-15 [?] CRAN (R 4.1.0)
P ggridges 0.5.3 2021-01-08 [?] CRAN (R 4.1.0)
P git2r 0.29.0 2021-11-22 [?] CRAN (R 4.1.0)
P glmGamPoi * 1.6.0 2021-10-26 [?] Bioconductor
P GlobalOptions 0.1.2 2020-06-10 [?] CRAN (R 4.1.0)
P globals 0.14.0 2020-11-22 [?] CRAN (R 4.0.2)
P globaltest 5.48.0 2021-10-26 [?] Bioconductor
P glue * 1.6.0 2021-12-17 [?] CRAN (R 4.1.0)
P GO.db * 3.14.0 2021-12-21 [?] Bioconductor
P goftest 1.2-3 2021-10-07 [?] CRAN (R 4.1.0)
P gplots 3.1.1 2020-11-28 [?] CRAN (R 4.0.2)
P graph * 1.72.0 2021-10-26 [?] Bioconductor
P graphlayouts 0.8.0 2022-01-03 [?] CRAN (R 4.1.0)
P gridExtra 2.3 2017-09-09 [?] CRAN (R 4.1.0)
P GSA 1.03.1 2019-01-31 [?] CRAN (R 4.1.0)
P GSEABase 1.56.0 2021-10-26 [?] Bioconductor
P GSVA 1.42.0 2021-10-26 [?] Bioconductor
P gtable 0.3.0 2019-03-25 [?] CRAN (R 4.1.0)
P gtools 3.9.2 2021-06-06 [?] CRAN (R 4.1.0)
P haven 2.4.3 2021-08-04 [?] CRAN (R 4.1.0)
P HDF5Array 1.22.1 2021-11-14 [?] Bioconductor
P here * 1.0.1 2020-12-13 [?] CRAN (R 4.0.2)
P hgu133a.db 3.13.0 2022-01-24 [?] Bioconductor
P hgu133plus2.db 3.13.0 2022-01-24 [?] Bioconductor
P highr 0.9 2021-04-16 [?] CRAN (R 4.1.0)
P hms 1.1.1 2021-09-26 [?] CRAN (R 4.1.0)
P htmltools 0.5.2 2021-08-25 [?] CRAN (R 4.1.0)
P HTMLUtils 0.1.7 2015-01-17 [?] CRAN (R 4.1.0)
P htmlwidgets 1.5.4 2021-09-08 [?] CRAN (R 4.1.0)
P httpuv 1.6.5 2022-01-05 [?] CRAN (R 4.1.0)
P httr 1.4.2 2020-07-20 [?] CRAN (R 4.1.0)
P hwriter 1.3.2 2014-09-10 [?] CRAN (R 4.1.0)
P ica 1.0-2 2018-05-24 [?] CRAN (R 4.1.0)
P igraph 1.2.11 2022-01-04 [?] CRAN (R 4.1.0)
P IRanges * 2.28.0 2021-10-26 [?] Bioconductor
P irlba 2.3.5 2021-12-06 [?] CRAN (R 4.1.0)
P iterators 1.0.13 2020-10-15 [?] CRAN (R 4.0.2)
P jquerylib 0.1.4 2021-04-26 [?] CRAN (R 4.1.0)
P jsonlite 1.7.2 2020-12-09 [?] CRAN (R 4.0.2)
P KEGGdzPathwaysGEO 1.32.0 2021-10-30 [?] Bioconductor
P KEGGgraph 1.54.0 2021-10-26 [?] Bioconductor
P KEGGREST 1.34.0 2021-10-26 [?] Bioconductor
P KernSmooth 2.23-20 2021-05-03 [?] CRAN (R 4.1.0)
P knitr 1.37 2021-12-16 [?] CRAN (R 4.1.0)
P labeling 0.4.2 2020-10-20 [?] CRAN (R 4.0.2)
P later 1.3.0 2021-08-18 [?] CRAN (R 4.1.0)
P lattice 0.20-45 2021-09-22 [?] CRAN (R 4.1.0)
P lazyeval 0.2.2 2019-03-15 [?] CRAN (R 4.1.0)
P leiden 0.3.9 2021-07-27 [?] CRAN (R 4.1.0)
P lifecycle 1.0.1 2021-09-24 [?] CRAN (R 4.1.0)
P limma * 3.50.0 2021-10-26 [?] Bioconductor
P listenv 0.8.0 2019-12-05 [?] CRAN (R 4.1.0)
P lmtest 0.9-39 2021-11-07 [?] CRAN (R 4.1.0)
P locfit 1.5-9.4 2020-03-25 [?] CRAN (R 4.1.0)
P lubridate 1.8.0 2021-10-07 [?] CRAN (R 4.1.0)
P magrittr 2.0.1 2020-11-17 [?] CRAN (R 4.0.2)
P MASS 7.3-53.1 2021-02-12 [?] CRAN (R 4.0.2)
P mathjaxr 1.4-0 2021-03-01 [?] CRAN (R 4.1.0)
P Matrix 1.4-0 2021-12-08 [?] CRAN (R 4.1.0)
P MatrixGenerics * 1.6.0 2021-10-26 [?] Bioconductor
P matrixStats * 0.61.0 2021-09-17 [?] CRAN (R 4.1.0)
P memoise 2.0.1 2021-11-26 [?] CRAN (R 4.1.0)
P metap 1.7 2021-12-16 [?] CRAN (R 4.1.0)
P metapod 1.2.0 2021-10-26 [?] Bioconductor
P mgcv 1.8-38 2021-10-06 [?] CRAN (R 4.1.0)
P mime 0.12 2021-09-28 [?] CRAN (R 4.1.0)
P miniUI 0.1.1.1 2018-05-18 [?] CRAN (R 4.1.0)
P mnormt 2.0.2 2020-09-01 [?] CRAN (R 4.0.2)
P modelr 0.1.8 2020-05-19 [?] CRAN (R 4.0.2)
P multcomp 1.4-18 2022-01-04 [?] CRAN (R 4.1.0)
P multtest 2.50.0 2021-10-26 [?] Bioconductor
P munsell 0.5.0 2018-06-12 [?] CRAN (R 4.1.0)
P mutoss 0.1-12 2017-12-04 [?] CRAN (R 4.1.0)
P mvtnorm 1.1-3 2021-10-08 [?] CRAN (R 4.1.0)
P nlme 3.1-153 2021-09-07 [?] CRAN (R 4.1.0)
P numDeriv 2016.8-1.1 2019-06-06 [?] CRAN (R 4.1.0)
P org.Hs.eg.db * 3.14.0 2021-12-21 [?] Bioconductor
P org.Mm.eg.db 3.14.0 2022-01-24 [?] Bioconductor
P org.Rn.eg.db 3.14.0 2022-01-24 [?] Bioconductor
P PADOG 1.36.0 2021-10-26 [?] Bioconductor
P paletteer * 1.4.0 2021-07-20 [?] CRAN (R 4.1.0)
P parallelly 1.30.0 2021-12-17 [?] CRAN (R 4.1.0)
P patchwork * 1.1.1 2020-12-17 [?] CRAN (R 4.0.2)
P pathview * 1.34.0 2021-10-26 [?] Bioconductor
P pbapply 1.5-0 2021-09-16 [?] CRAN (R 4.1.0)
P pillar 1.6.4 2021-10-18 [?] CRAN (R 4.1.0)
P pkgconfig 2.0.3 2019-09-22 [?] CRAN (R 4.1.0)
P plotly 4.10.0 2021-10-09 [?] CRAN (R 4.1.0)
P plotrix 3.8-2 2021-09-08 [?] CRAN (R 4.1.0)
P plyr 1.8.6 2020-03-03 [?] CRAN (R 4.0.2)
P png 0.1-7 2013-12-03 [?] CRAN (R 4.1.0)
P polyclip 1.10-0 2019-03-14 [?] CRAN (R 4.1.0)
P processx 3.5.2 2021-04-30 [?] CRAN (R 4.1.0)
P promises 1.2.0.1 2021-02-11 [?] CRAN (R 4.0.2)
P ps 1.6.0 2021-02-28 [?] CRAN (R 4.1.0)
P purrr * 0.3.4 2020-04-17 [?] CRAN (R 4.0.2)
P R.methodsS3 1.8.1 2020-08-26 [?] CRAN (R 4.0.2)
P R.oo 1.24.0 2020-08-26 [?] CRAN (R 4.0.2)
P R.utils 2.11.0 2021-09-26 [?] CRAN (R 4.1.0)
P R2HTML 2.3.2 2016-06-23 [?] CRAN (R 4.1.0)
P R6 2.5.1 2021-08-19 [?] CRAN (R 4.1.0)
P RANN 2.6.1 2019-01-08 [?] CRAN (R 4.1.0)
P rbibutils 2.2.7 2021-12-07 [?] CRAN (R 4.1.0)
P RColorBrewer 1.1-2 2014-12-07 [?] CRAN (R 4.0.2)
P Rcpp 1.0.7 2021-07-07 [?] CRAN (R 4.1.0)
P RcppAnnoy 0.0.19 2021-07-30 [?] CRAN (R 4.1.0)
RCurl 1.98-1.6 2022-02-08 [1] CRAN (R 4.1.0)
P Rdpack 2.1.3 2021-12-08 [?] CRAN (R 4.1.0)
P readr * 2.1.1 2021-11-30 [?] CRAN (R 4.1.0)
P readxl 1.3.1 2019-03-13 [?] CRAN (R 4.1.0)
P rematch2 2.1.2 2020-05-01 [?] CRAN (R 4.1.0)
P renv 0.15.0-14 2022-01-10 [?] Github (rstudio/renv@a3b90eb)
P reprex 2.0.1 2021-08-05 [?] CRAN (R 4.1.0)
P reshape2 1.4.4 2020-04-09 [?] CRAN (R 4.1.0)
P reticulate 1.22 2021-09-17 [?] CRAN (R 4.1.0)
P Rgraphviz 2.38.0 2021-10-26 [?] Bioconductor
P rhdf5 2.38.0 2021-10-26 [?] Bioconductor
P rhdf5filters 1.6.0 2021-10-26 [?] Bioconductor
P Rhdf5lib 1.16.0 2021-10-26 [?] Bioconductor
P rjson 0.2.21 2022-01-09 [?] CRAN (R 4.1.0)
P rlang 0.4.12 2021-10-18 [?] CRAN (R 4.1.0)
P rmarkdown 2.11 2021-09-14 [?] CRAN (R 4.1.0)
P rngtools 1.5.2 2021-09-20 [?] CRAN (R 4.1.0)
P ROCR 1.0-11 2020-05-02 [?] CRAN (R 4.1.0)
P rpart 4.1-15 2019-04-12 [?] CRAN (R 4.1.0)
P rprojroot 2.0.2 2020-11-15 [?] CRAN (R 4.0.2)
P RSpectra 0.16-0 2019-12-01 [?] CRAN (R 4.1.0)
P RSQLite 2.2.9 2021-12-06 [?] CRAN (R 4.1.0)
P rstudioapi 0.13 2020-11-12 [?] CRAN (R 4.0.2)
P rsvd 1.0.5 2021-04-16 [?] CRAN (R 4.1.0)
P Rtsne 0.15 2018-11-10 [?] CRAN (R 4.1.0)
P rvest 1.0.2 2021-10-16 [?] CRAN (R 4.1.0)
P S4Vectors * 0.32.3 2021-11-21 [?] Bioconductor
P safe 3.34.0 2021-10-26 [?] Bioconductor
P sandwich 3.0-1 2021-05-18 [?] CRAN (R 4.1.0)
P sass 0.4.0 2021-05-12 [?] CRAN (R 4.1.0)
P ScaledMatrix 1.2.0 2021-10-26 [?] Bioconductor
P scales 1.1.1 2020-05-11 [?] CRAN (R 4.0.2)
P scater * 1.22.0 2021-10-26 [?] Bioconductor
P scattermore 0.7 2020-11-24 [?] CRAN (R 4.1.0)
P scran * 1.22.1 2021-11-14 [?] Bioconductor
P sctransform 0.3.3 2022-01-13 [?] CRAN (R 4.1.0)
P scuttle * 1.4.0 2021-10-26 [?] Bioconductor
P sessioninfo 1.2.2 2021-12-06 [?] CRAN (R 4.1.0)
P Seurat * 4.0.6 2021-12-16 [?] CRAN (R 4.1.0)
P SeuratObject * 4.0.4 2021-11-23 [?] CRAN (R 4.1.0)
P shape 1.4.6 2021-05-19 [?] CRAN (R 4.1.0)
P shiny 1.7.1 2021-10-02 [?] CRAN (R 4.1.0)
P SingleCellExperiment * 1.16.0 2021-10-26 [?] Bioconductor
P sn 2.0.1 2021-11-26 [?] CRAN (R 4.1.0)
P SparseM * 1.81 2021-02-18 [?] CRAN (R 4.1.0)
P sparseMatrixStats 1.6.0 2021-10-26 [?] Bioconductor
P spatstat.core 2.3-2 2021-11-26 [?] CRAN (R 4.1.0)
P spatstat.data 2.1-2 2021-12-17 [?] CRAN (R 4.1.0)
P spatstat.geom 2.3-1 2021-12-10 [?] CRAN (R 4.1.0)
P spatstat.sparse 2.1-0 2021-12-17 [?] CRAN (R 4.1.0)
P spatstat.utils 2.3-0 2021-12-12 [?] CRAN (R 4.1.0)
P statmod 1.4.36 2021-05-10 [?] CRAN (R 4.1.0)
P stringi 1.7.6 2021-11-29 [?] CRAN (R 4.1.0)
P stringr * 1.4.0 2019-02-10 [?] CRAN (R 4.0.2)
P SummarizedExperiment * 1.24.0 2021-10-26 [?] Bioconductor
P survival 3.2-13 2021-08-24 [?] CRAN (R 4.1.0)
P tensor 1.5 2012-05-05 [?] CRAN (R 4.1.0)
P TFisher 0.2.0 2018-03-21 [?] CRAN (R 4.1.0)
P TH.data 1.1-0 2021-09-27 [?] CRAN (R 4.1.0)
P tibble * 3.1.6 2021-11-07 [?] CRAN (R 4.1.0)
P tidygraph 1.2.0 2020-05-12 [?] CRAN (R 4.0.2)
P tidyHeatmap * 1.7.0 2022-05-13 [?] Github (stemangiola/tidyHeatmap@241aec2)
P tidyr * 1.1.4 2021-09-27 [?] CRAN (R 4.1.0)
P tidyselect 1.1.1 2021-04-30 [?] CRAN (R 4.1.0)
P tidyverse * 1.3.1 2021-04-15 [?] CRAN (R 4.1.0)
P tmvnsim 1.0-2 2016-12-15 [?] CRAN (R 4.1.0)
P topGO * 2.46.0 2021-10-26 [?] Bioconductor
P tweenr 1.0.2 2021-03-23 [?] CRAN (R 4.1.0)
P tzdb 0.2.0 2021-10-27 [?] CRAN (R 4.1.0)
P utf8 1.2.2 2021-07-24 [?] CRAN (R 4.1.0)
P uwot 0.1.11 2021-12-02 [?] CRAN (R 4.1.0)
P vctrs 0.3.8 2021-04-29 [?] CRAN (R 4.0.2)
P vipor 0.4.5 2017-03-22 [?] CRAN (R 4.1.0)
P viridis 0.6.2 2021-10-13 [?] CRAN (R 4.1.0)
P viridisLite 0.4.0 2021-04-13 [?] CRAN (R 4.0.2)
P vroom 1.5.7 2021-11-30 [?] CRAN (R 4.1.0)
P whisker 0.4 2019-08-28 [?] CRAN (R 4.0.2)
P withr 2.4.3 2021-11-30 [?] CRAN (R 4.1.0)
P workflowr * 1.7.0 2021-12-21 [?] CRAN (R 4.1.0)
P xfun 0.29 2021-12-14 [?] CRAN (R 4.1.0)
P XML 3.99-0.8 2021-09-17 [?] CRAN (R 4.1.0)
P xml2 1.3.3 2021-11-30 [?] CRAN (R 4.1.0)
P xtable 1.8-4 2019-04-21 [?] CRAN (R 4.1.0)
P XVector 0.34.0 2021-10-26 [?] Bioconductor
P yaml 2.2.1 2020-02-01 [?] CRAN (R 4.0.2)
P zlibbioc 1.40.0 2021-10-26 [?] Bioconductor
P zoo 1.8-9 2021-03-09 [?] CRAN (R 4.1.0)
[1] /oshlack_lab/jovana.maksimovic/projects/MCRI/melanie.neeland/paed-cf-cite-seq/renv/library/R-4.1/x86_64-pc-linux-gnu
[2] /config/binaries/R/4.1.0/lib64/R/library
P ── Loaded and on-disk path mismatch.
──────────────────────────────────────────────────────────────────────────────9 References
sessionInfo()R version 4.1.0 (2021-05-18)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: CentOS Linux 7 (Core)
Matrix products: default
BLAS: /config/binaries/R/4.1.0/lib64/R/lib/libRblas.so
LAPACK: /config/binaries/R/4.1.0/lib64/R/lib/libRlapack.so
locale:
[1] LC_CTYPE=en_AU.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_AU.UTF-8 LC_COLLATE=en_AU.UTF-8
[5] LC_MONETARY=en_AU.UTF-8 LC_MESSAGES=en_AU.UTF-8
[7] LC_PAPER=en_AU.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_AU.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats4 stats graphics grDevices datasets utils methods
[8] base
other attached packages:
[1] EGSEA_1.22.0 pathview_1.34.0
[3] topGO_2.46.0 SparseM_1.81
[5] GO.db_3.14.0 graph_1.72.0
[7] gage_2.44.0 org.Hs.eg.db_3.14.0
[9] AnnotationDbi_1.56.2 edgeR_3.36.0
[11] limma_3.50.0 tidyHeatmap_1.7.0
[13] paletteer_1.4.0 BiocParallel_1.28.3
[15] glmGamPoi_1.6.0 clustree_0.4.4
[17] ggraph_2.0.5 patchwork_1.1.1
[19] SeuratObject_4.0.4 Seurat_4.0.6
[21] scater_1.22.0 scran_1.22.1
[23] scuttle_1.4.0 DropletUtils_1.14.1
[25] SingleCellExperiment_1.16.0 SummarizedExperiment_1.24.0
[27] Biobase_2.54.0 GenomicRanges_1.46.1
[29] GenomeInfoDb_1.30.1 IRanges_2.28.0
[31] S4Vectors_0.32.3 BiocGenerics_0.40.0
[33] MatrixGenerics_1.6.0 matrixStats_0.61.0
[35] glue_1.6.0 here_1.0.1
[37] forcats_0.5.1 stringr_1.4.0
[39] dplyr_1.0.7 purrr_0.3.4
[41] readr_2.1.1 tidyr_1.1.4
[43] tibble_3.1.6 ggplot2_3.3.5
[45] tidyverse_1.3.1 BiocStyle_2.22.0
[47] workflowr_1.7.0
loaded via a namespace (and not attached):
[1] rsvd_1.0.5 ica_1.0-2
[3] ps_1.6.0 foreach_1.5.1
[5] lmtest_0.9-39 rprojroot_2.0.2
[7] crayon_1.4.2 rbibutils_2.2.7
[9] spatstat.core_2.3-2 MASS_7.3-53.1
[11] rhdf5filters_1.6.0 nlme_3.1-153
[13] backports_1.4.1 reprex_2.0.1
[15] rlang_0.4.12 XVector_0.34.0
[17] ROCR_1.0-11 readxl_1.3.1
[19] irlba_2.3.5 callr_3.7.0
[21] rjson_0.2.21 globaltest_5.48.0
[23] bit64_4.0.5 rngtools_1.5.2
[25] sctransform_0.3.3 parallel_4.1.0
[27] processx_3.5.2 vipor_0.4.5
[29] spatstat.sparse_2.1-0 R2HTML_2.3.2
[31] spatstat.geom_2.3-1 haven_2.4.3
[33] tidyselect_1.1.1 fitdistrplus_1.1-6
[35] XML_3.99-0.8 zoo_1.8-9
[37] org.Mm.eg.db_3.14.0 xtable_1.8-4
[39] magrittr_2.0.1 evaluate_0.14
[41] Rdpack_2.1.3 cli_3.1.0
[43] zlibbioc_1.40.0 sn_2.0.1
[45] hwriter_1.3.2 doRNG_1.8.2
[47] rstudioapi_0.13 miniUI_0.1.1.1
[49] whisker_0.4 bslib_0.3.1
[51] rpart_4.1-15 mathjaxr_1.4-0
[53] GSA_1.03.1 KEGGdzPathwaysGEO_1.32.0
[55] shiny_1.7.1 GSVA_1.42.0
[57] BiocSingular_1.10.0 xfun_0.29
[59] clue_0.3-60 org.Rn.eg.db_3.14.0
[61] multtest_2.50.0 cluster_2.1.2
[63] caTools_1.18.2 tidygraph_1.2.0
[65] KEGGREST_1.34.0 ggrepel_0.9.1
[67] listenv_0.8.0 dendextend_1.15.2
[69] Biostrings_2.62.0 png_0.1-7
[71] future_1.23.0 withr_2.4.3
[73] bitops_1.0-7 ggforce_0.3.3
[75] plyr_1.8.6 cellranger_1.1.0
[77] PADOG_1.36.0 GSEABase_1.56.0
[79] dqrng_0.3.0 pillar_1.6.4
[81] gplots_3.1.1 GlobalOptions_0.1.2
[83] cachem_1.0.6 multcomp_1.4-18
[85] fs_1.5.2 GetoptLong_1.0.5
[87] DelayedMatrixStats_1.16.0 vctrs_0.3.8
[89] ellipsis_0.3.2 generics_0.1.1
[91] metap_1.7 tools_4.1.0
[93] beeswarm_0.4.0 munsell_0.5.0
[95] tweenr_1.0.2 DelayedArray_0.20.0
[97] fastmap_1.1.0 compiler_4.1.0
[99] abind_1.4-5 httpuv_1.6.5
[101] sessioninfo_1.2.2 plotly_4.10.0
[103] GenomeInfoDbData_1.2.7 gridExtra_2.3
[105] lattice_0.20-45 deldir_1.0-6
[107] mutoss_0.1-12 utf8_1.2.2
[109] later_1.3.0 jsonlite_1.7.2
[111] scales_1.1.1 ScaledMatrix_1.2.0
[113] pbapply_1.5-0 sparseMatrixStats_1.6.0
[115] renv_0.15.0-14 lazyeval_0.2.2
[117] promises_1.2.0.1 doParallel_1.0.16
[119] R.utils_2.11.0 goftest_1.2-3
[121] checkmate_2.0.0 spatstat.utils_2.3-0
[123] reticulate_1.22 sandwich_3.0-1
[125] rmarkdown_2.11 cowplot_1.1.1
[127] statmod_1.4.36 Rtsne_0.15
[129] EGSEAdata_1.22.0 uwot_0.1.11
[131] igraph_1.2.11 HDF5Array_1.22.1
[133] plotrix_3.8-2 numDeriv_2016.8-1.1
[135] survival_3.2-13 yaml_2.2.1
[137] htmltools_0.5.2 memoise_2.0.1
[139] locfit_1.5-9.4 graphlayouts_0.8.0
[141] viridisLite_0.4.0 digest_0.6.29
[143] assertthat_0.2.1 mime_0.12
[145] RSQLite_2.2.9 future.apply_1.8.1
[147] data.table_1.14.2 blob_1.2.2
[149] R.oo_1.24.0 labeling_0.4.2
[151] splines_4.1.0 rematch2_2.1.2
[153] Rhdf5lib_1.16.0 RCurl_1.98-1.6
[155] broom_0.7.11 hms_1.1.1
[157] modelr_0.1.8 rhdf5_2.38.0
[159] colorspace_2.0-2 mnormt_2.0.2
[161] BiocManager_1.30.16 tmvnsim_1.0-2
[163] ggbeeswarm_0.6.0 shape_1.4.6
[165] sass_0.4.0 Rcpp_1.0.7
[167] bookdown_0.24 RANN_2.6.1
[169] mvtnorm_1.1-3 circlize_0.4.13
[171] fansi_1.0.0 tzdb_0.2.0
[173] parallelly_1.30.0 R6_2.5.1
[175] grid_4.1.0 ggridges_0.5.3
[177] lifecycle_1.0.1 TFisher_0.2.0
[179] bluster_1.4.0 leiden_0.3.9
[181] jquerylib_0.1.4 safe_3.34.0
[183] Matrix_1.4-0 TH.data_1.1-0
[185] RcppAnnoy_0.0.19 RColorBrewer_1.1-2
[187] iterators_1.0.13 htmlwidgets_1.5.4
[189] beachmat_2.10.0 polyclip_1.10-0
[191] rvest_1.0.2 ComplexHeatmap_2.10.0
[193] mgcv_1.8-38 globals_0.14.0
[195] hgu133plus2.db_3.13.0 KEGGgraph_1.54.0
[197] codetools_0.2-18 lubridate_1.8.0
[199] metapod_1.2.0 gtools_3.9.2
[201] getPass_0.2-2 dbplyr_2.1.1
[203] RSpectra_0.16-0 R.methodsS3_1.8.1
[205] gtable_0.3.0 DBI_1.1.2
[207] git2r_0.29.0 highr_0.9
[209] tensor_1.5 httr_1.4.2
[211] KernSmooth_2.23-20 vroom_1.5.7
[213] stringi_1.7.6 reshape2_1.4.4
[215] farver_2.1.0 annotate_1.72.0
[217] viridis_0.6.2 Rgraphviz_2.38.0
[219] DT_0.20 xml2_1.3.3
[221] BiocNeighbors_1.12.0 scattermore_0.7
[223] bit_4.0.4 spatstat.data_2.1-2
[225] hgu133a.db_3.13.0 pkgconfig_2.0.3
[227] HTMLUtils_0.1.7 knitr_1.37