Remove ambient RNA contamination
Run decontX
During this step we will also denoise the ADT data for each batch
using DSB as outlined in this workflow .
# identify isotype controls for DSB ADT normalisation
read_csv(file = here("data",
"C133_Neeland_batch1",
"data",
"sample_sheets",
"ADT_features.csv")) %>%
dplyr::filter(grepl("[Ii]sotype", id)) %>%
pull(name) -> isotype_controls
sceLst <- lapply(1:length(sceLst), function(i){
sce <- sceLst[[i]]
sce_raw <- readRDS(str_replace(names(sceLst)[i],
"doublets_filtered",
"CellRanger"))
if(length(levels(sce$Capture)) < 4){
sce_decont <- decontX(sce, background = sce_raw)
# get isotype controls
rowData(altExp(sce, "ADT")) %>%
data.frame %>%
dplyr::filter(grepl("[Ii]sotype", Symbol)) %>%
pull(ID) -> isotype_controls
# get ADT counts for "cells"
adt <- counts(altExp(sce, "ADT"))
# get ADT counts for background
adt_background <- counts(altExp(sce_raw, "Antibody Capture"))
# exclude all "cells" from background matrix
adt_background <- adt_background[!str_detect(rownames(adt_background), "HTO"),]
adt_background <- adt_background[,!colnames(adt_background) %in% colnames(adt)]
# exclude droplets with >500 RNA counts
adt_background <- adt_background[, colSums(adt_background) < 500]
# normalize and denoise with dsb
adt_dsb <- DSBNormalizeProtein(
cell_protein_matrix = adt,
empty_drop_matrix = adt_background,
denoise.counts = TRUE,
use.isotype.control = TRUE,
isotype.control.name.vec = isotype_controls)
# add normalised dsb ADT assay
tmp <- SingleCellExperiment(list(counts = adt_dsb),
rowData = rowData(altExp(sce_decont, "ADT")))
altExp(sce_decont, "ADT.dsb") <- tmp
} else {
sce_decont <- decontX(sce, background = sce_raw,
batch = sce$Capture,
bgBatch = sce_raw$Sample)
}
sce_decont
})[1] "correcting ambient protein background noise"
[1] "some proteins with low background variance detected check raw and normalized distributions. protein stats can be returned with return.stats = TRUE"
[1] "A0006" "A0007" "A0020" "A0023" "A0024" "A0026" "A0029" "A0031" "A0032"
[10] "A0033" "A0034" "A0046" "A0047" "A0050" "A0052" "A0053" "A0058" "A0063"
[19] "A0064" "A0066" "A0070" "A0071" "A0072" "A0073" "A0081" "A0083" "A0085"
[28] "A0087" "A0089" "A0090" "A0091" "A0092" "A0095" "A0100" "A0101" "A0124"
[37] "A0127" "A0134" "A0138" "A0140" "A0141" "A0142" "A0143" "A0144" "A0145"
[46] "A0146" "A0147" "A0149" "A0151" "A0152" "A0153" "A0154" "A0155" "A0156"
[55] "A0159" "A0160" "A0161" "A0162" "A0165" "A0167" "A0168" "A0170" "A0171"
[64] "A0172" "A0174" "A0176" "A0180" "A0181" "A0185" "A0187" "A0189" "A0206"
[73] "A0214" "A0215" "A0216" "A0217" "A0218" "A0219" "A0224" "A0236" "A0237"
[82] "A0238" "A0240" "A0241" "A0242" "A0246" "A0247" "A0352" "A0353" "A0355"
[91] "A0357" "A0358" "A0359" "A0364" "A0367" "A0368" "A0369" "A0370" "A0371"
[100] "A0372" "A0383" "A0384" "A0385" "A0386" "A0389" "A0390" "A0391" "A0393"
[109] "A0396" "A0398" "A0404" "A0406" "A0407" "A0408" "A0419" "A0420" "A0446"
[118] "A0575" "A0576" "A0577" "A0579" "A0581" "A0582" "A0586" "A0590" "A0591"
[127] "A0599" "A0817" "A0822" "A0830" "A0845" "A0853" "A0861" "A0864" "A0866"
[136] "A0867" "A0868" "A0871" "A0872" "A0896" "A0902" "A0912" "A0920" "A0923"
[145] "A0931" "A0940" "A0941" "A0944" "A1018" "A1046"
[1] "fitting models to each cell for dsb technical component and removing cell to cell technical noise"
[1] "correcting ambient protein background noise"
[1] "some proteins with low background variance detected check raw and normalized distributions. protein stats can be returned with return.stats = TRUE"
[1] "A0006" "A0007" "A0020" "A0023" "A0024" "A0026" "A0029" "A0031" "A0032"
[10] "A0033" "A0034" "A0046" "A0047" "A0050" "A0052" "A0053" "A0058" "A0063"
[19] "A0064" "A0066" "A0070" "A0071" "A0072" "A0073" "A0081" "A0083" "A0085"
[28] "A0087" "A0088" "A0089" "A0090" "A0091" "A0092" "A0095" "A0100" "A0101"
[37] "A0124" "A0127" "A0134" "A0138" "A0140" "A0141" "A0142" "A0143" "A0144"
[46] "A0145" "A0146" "A0147" "A0149" "A0151" "A0152" "A0153" "A0154" "A0155"
[55] "A0156" "A0158" "A0159" "A0160" "A0161" "A0162" "A0163" "A0165" "A0167"
[64] "A0168" "A0170" "A0171" "A0172" "A0174" "A0176" "A0179" "A0180" "A0181"
[73] "A0185" "A0187" "A0189" "A0206" "A0214" "A0215" "A0216" "A0217" "A0218"
[82] "A0219" "A0224" "A0236" "A0237" "A0238" "A0240" "A0241" "A0242" "A0246"
[91] "A0247" "A0352" "A0353" "A0355" "A0357" "A0358" "A0359" "A0364" "A0367"
[100] "A0368" "A0369" "A0370" "A0371" "A0372" "A0373" "A0383" "A0384" "A0385"
[109] "A0386" "A0389" "A0390" "A0391" "A0393" "A0396" "A0398" "A0404" "A0406"
[118] "A0407" "A0408" "A0419" "A0420" "A0446" "A0447" "A0575" "A0576" "A0577"
[127] "A0579" "A0581" "A0582" "A0586" "A0590" "A0591" "A0599" "A0817" "A0822"
[136] "A0830" "A0845" "A0853" "A0861" "A0864" "A0866" "A0867" "A0868" "A0870"
[145] "A0871" "A0872" "A0894" "A0896" "A0897" "A0898" "A0902" "A0912" "A0920"
[154] "A0923" "A0931" "A0935" "A0940" "A0941" "A0944" "A1018" "A1046"
[1] "fitting models to each cell for dsb technical component and removing cell to cell technical noise"
[1] "correcting ambient protein background noise"
[1] "some proteins with low background variance detected check raw and normalized distributions. protein stats can be returned with return.stats = TRUE"
[1] "A0006" "A0007" "A0020" "A0023" "A0024" "A0026" "A0029" "A0031" "A0032"
[10] "A0033" "A0034" "A0046" "A0047" "A0050" "A0052" "A0053" "A0058" "A0063"
[19] "A0064" "A0066" "A0070" "A0071" "A0072" "A0073" "A0081" "A0083" "A0085"
[28] "A0087" "A0088" "A0089" "A0090" "A0091" "A0092" "A0095" "A0100" "A0101"
[37] "A0124" "A0127" "A0134" "A0136" "A0138" "A0140" "A0141" "A0142" "A0143"
[46] "A0144" "A0145" "A0146" "A0147" "A0149" "A0151" "A0152" "A0153" "A0154"
[55] "A0155" "A0156" "A0158" "A0159" "A0160" "A0161" "A0162" "A0163" "A0165"
[64] "A0167" "A0168" "A0170" "A0171" "A0172" "A0174" "A0176" "A0179" "A0180"
[73] "A0181" "A0185" "A0187" "A0189" "A0206" "A0214" "A0215" "A0216" "A0217"
[82] "A0218" "A0219" "A0224" "A0236" "A0237" "A0238" "A0240" "A0241" "A0242"
[91] "A0246" "A0247" "A0352" "A0353" "A0355" "A0357" "A0358" "A0359" "A0364"
[100] "A0367" "A0368" "A0369" "A0370" "A0371" "A0372" "A0373" "A0383" "A0384"
[109] "A0385" "A0386" "A0389" "A0390" "A0391" "A0393" "A0394" "A0396" "A0398"
[118] "A0404" "A0406" "A0407" "A0408" "A0419" "A0420" "A0446" "A0447" "A0575"
[127] "A0576" "A0577" "A0579" "A0581" "A0582" "A0586" "A0590" "A0591" "A0599"
[136] "A0817" "A0822" "A0830" "A0845" "A0853" "A0861" "A0864" "A0866" "A0867"
[145] "A0868" "A0870" "A0871" "A0872" "A0894" "A0896" "A0897" "A0898" "A0902"
[154] "A0912" "A0920" "A0923" "A0931" "A0935" "A0940" "A0941" "A0944" "A1018"
[163] "A1046"
[1] "fitting models to each cell for dsb technical component and removing cell to cell technical noise"
[1] "correcting ambient protein background noise"
[1] "some proteins with low background variance detected check raw and normalized distributions. protein stats can be returned with return.stats = TRUE"
[1] "A0006" "A0007" "A0020" "A0023" "A0024" "A0026" "A0029" "A0031" "A0032"
[10] "A0033" "A0034" "A0046" "A0047" "A0050" "A0052" "A0053" "A0058" "A0063"
[19] "A0064" "A0066" "A0070" "A0071" "A0072" "A0073" "A0081" "A0083" "A0085"
[28] "A0087" "A0088" "A0089" "A0090" "A0091" "A0092" "A0095" "A0100" "A0101"
[37] "A0124" "A0127" "A0134" "A0136" "A0138" "A0140" "A0141" "A0142" "A0143"
[46] "A0144" "A0145" "A0146" "A0147" "A0149" "A0151" "A0152" "A0153" "A0154"
[55] "A0155" "A0156" "A0158" "A0159" "A0160" "A0161" "A0162" "A0163" "A0165"
[64] "A0167" "A0168" "A0170" "A0171" "A0172" "A0174" "A0176" "A0179" "A0180"
[73] "A0181" "A0185" "A0187" "A0189" "A0206" "A0214" "A0215" "A0216" "A0217"
[82] "A0218" "A0219" "A0224" "A0236" "A0237" "A0238" "A0240" "A0241" "A0242"
[91] "A0246" "A0247" "A0352" "A0353" "A0355" "A0357" "A0358" "A0359" "A0364"
[100] "A0367" "A0368" "A0369" "A0370" "A0371" "A0372" "A0373" "A0383" "A0384"
[109] "A0385" "A0386" "A0389" "A0390" "A0391" "A0393" "A0394" "A0396" "A0398"
[118] "A0404" "A0406" "A0407" "A0408" "A0419" "A0420" "A0446" "A0447" "A0575"
[127] "A0576" "A0577" "A0579" "A0581" "A0582" "A0586" "A0590" "A0591" "A0599"
[136] "A0817" "A0822" "A0830" "A0845" "A0853" "A0861" "A0864" "A0866" "A0867"
[145] "A0868" "A0870" "A0871" "A0872" "A0894" "A0896" "A0897" "A0898" "A0902"
[154] "A0912" "A0920" "A0923" "A0931" "A0935" "A0940" "A0941" "A0944" "A1018"
[163] "A1046"
[1] "fitting models to each cell for dsb technical component and removing cell to cell technical noise"
[1] "correcting ambient protein background noise"
[1] "some proteins with low background variance detected check raw and normalized distributions. protein stats can be returned with return.stats = TRUE"
[1] "A0006" "A0007" "A0020" "A0023" "A0024" "A0026" "A0029" "A0031" "A0032"
[10] "A0033" "A0034" "A0046" "A0047" "A0050" "A0052" "A0053" "A0058" "A0063"
[19] "A0064" "A0066" "A0070" "A0071" "A0072" "A0073" "A0081" "A0083" "A0085"
[28] "A0087" "A0088" "A0089" "A0090" "A0091" "A0092" "A0095" "A0100" "A0101"
[37] "A0124" "A0127" "A0134" "A0136" "A0138" "A0140" "A0141" "A0142" "A0143"
[46] "A0144" "A0145" "A0146" "A0147" "A0149" "A0151" "A0152" "A0153" "A0154"
[55] "A0155" "A0156" "A0158" "A0159" "A0160" "A0161" "A0162" "A0163" "A0165"
[64] "A0167" "A0168" "A0170" "A0171" "A0172" "A0174" "A0176" "A0179" "A0180"
[73] "A0181" "A0185" "A0187" "A0189" "A0206" "A0214" "A0215" "A0216" "A0217"
[82] "A0218" "A0219" "A0224" "A0236" "A0237" "A0238" "A0240" "A0241" "A0242"
[91] "A0246" "A0247" "A0352" "A0353" "A0355" "A0357" "A0358" "A0359" "A0364"
[100] "A0367" "A0368" "A0369" "A0370" "A0371" "A0372" "A0373" "A0383" "A0384"
[109] "A0385" "A0386" "A0389" "A0390" "A0391" "A0393" "A0394" "A0396" "A0398"
[118] "A0404" "A0406" "A0407" "A0408" "A0419" "A0420" "A0446" "A0447" "A0575"
[127] "A0576" "A0577" "A0579" "A0581" "A0582" "A0586" "A0590" "A0591" "A0599"
[136] "A0817" "A0822" "A0830" "A0845" "A0853" "A0861" "A0864" "A0866" "A0867"
[145] "A0868" "A0870" "A0871" "A0872" "A0894" "A0896" "A0897" "A0898" "A0902"
[154] "A0912" "A0920" "A0923" "A0931" "A0935" "A0940" "A0941" "A0944" "A1018"
[163] "A1046"
[1] "fitting models to each cell for dsb technical component and removing cell to cell technical noise"
[1] "correcting ambient protein background noise"
[1] "some proteins with low background variance detected check raw and normalized distributions. protein stats can be returned with return.stats = TRUE"
[1] "A0006" "A0007" "A0020" "A0023" "A0024" "A0026" "A0029" "A0031" "A0032"
[10] "A0033" "A0034" "A0046" "A0047" "A0050" "A0052" "A0053" "A0058" "A0063"
[19] "A0064" "A0066" "A0070" "A0071" "A0072" "A0073" "A0081" "A0083" "A0085"
[28] "A0087" "A0088" "A0089" "A0090" "A0091" "A0092" "A0095" "A0100" "A0101"
[37] "A0124" "A0127" "A0134" "A0136" "A0138" "A0140" "A0141" "A0142" "A0143"
[46] "A0144" "A0145" "A0146" "A0147" "A0149" "A0151" "A0152" "A0153" "A0154"
[55] "A0155" "A0156" "A0158" "A0159" "A0160" "A0161" "A0162" "A0163" "A0165"
[64] "A0167" "A0168" "A0170" "A0171" "A0172" "A0174" "A0176" "A0179" "A0180"
[73] "A0181" "A0185" "A0187" "A0189" "A0206" "A0214" "A0215" "A0216" "A0217"
[82] "A0218" "A0219" "A0224" "A0236" "A0237" "A0238" "A0240" "A0241" "A0242"
[91] "A0246" "A0247" "A0352" "A0353" "A0355" "A0357" "A0358" "A0359" "A0364"
[100] "A0367" "A0368" "A0369" "A0370" "A0371" "A0372" "A0373" "A0383" "A0384"
[109] "A0385" "A0386" "A0389" "A0390" "A0391" "A0393" "A0394" "A0396" "A0398"
[118] "A0404" "A0406" "A0407" "A0408" "A0419" "A0420" "A0446" "A0447" "A0575"
[127] "A0576" "A0577" "A0579" "A0581" "A0582" "A0586" "A0590" "A0591" "A0599"
[136] "A0817" "A0822" "A0830" "A0845" "A0853" "A0861" "A0864" "A0866" "A0867"
[145] "A0868" "A0870" "A0871" "A0872" "A0894" "A0896" "A0897" "A0898" "A0902"
[154] "A0912" "A0920" "A0923" "A0931" "A0935" "A0940" "A0941" "A0944" "A1018"
[163] "A1046"
[1] "fitting models to each cell for dsb technical component and removing cell to cell technical noise"
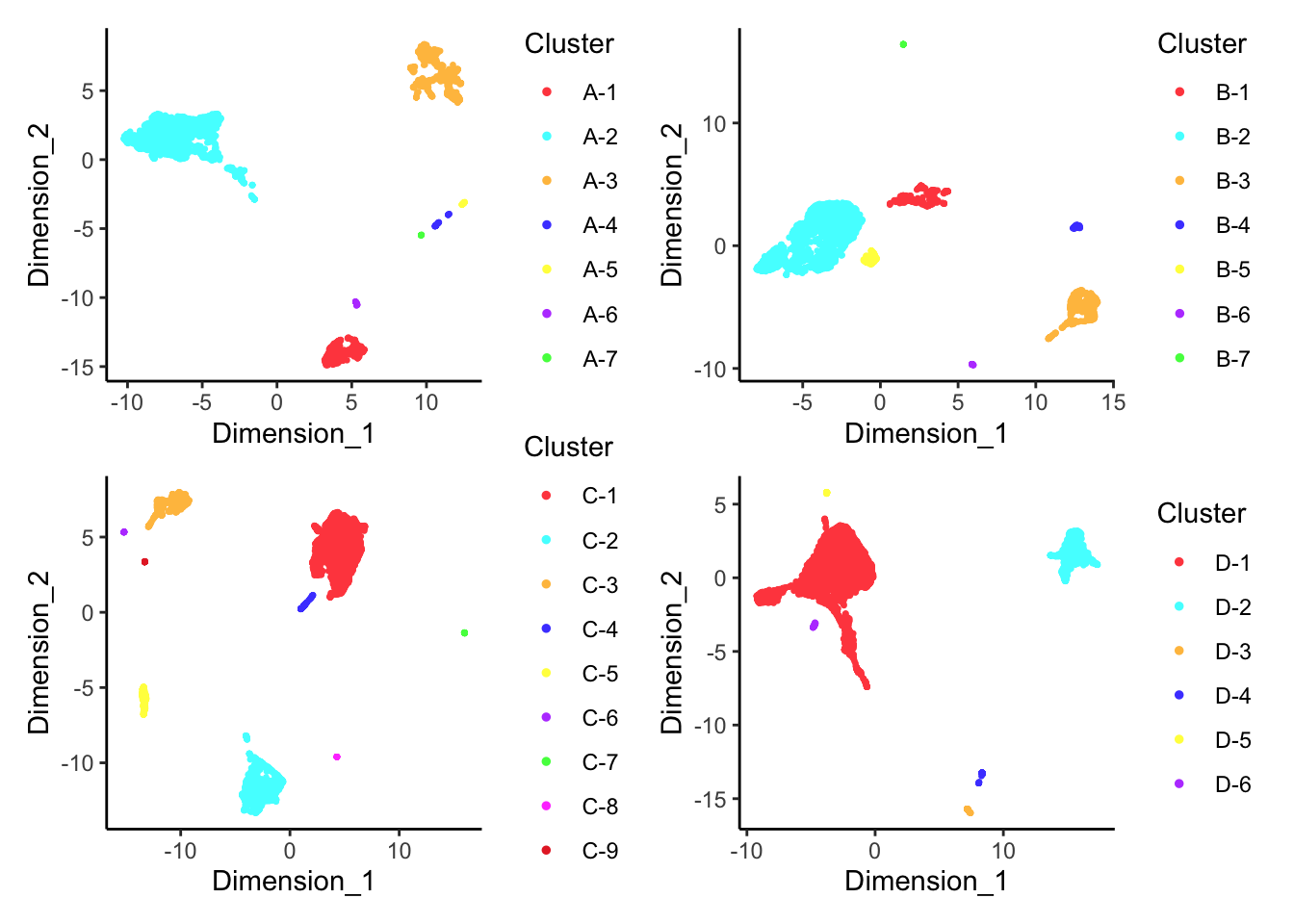
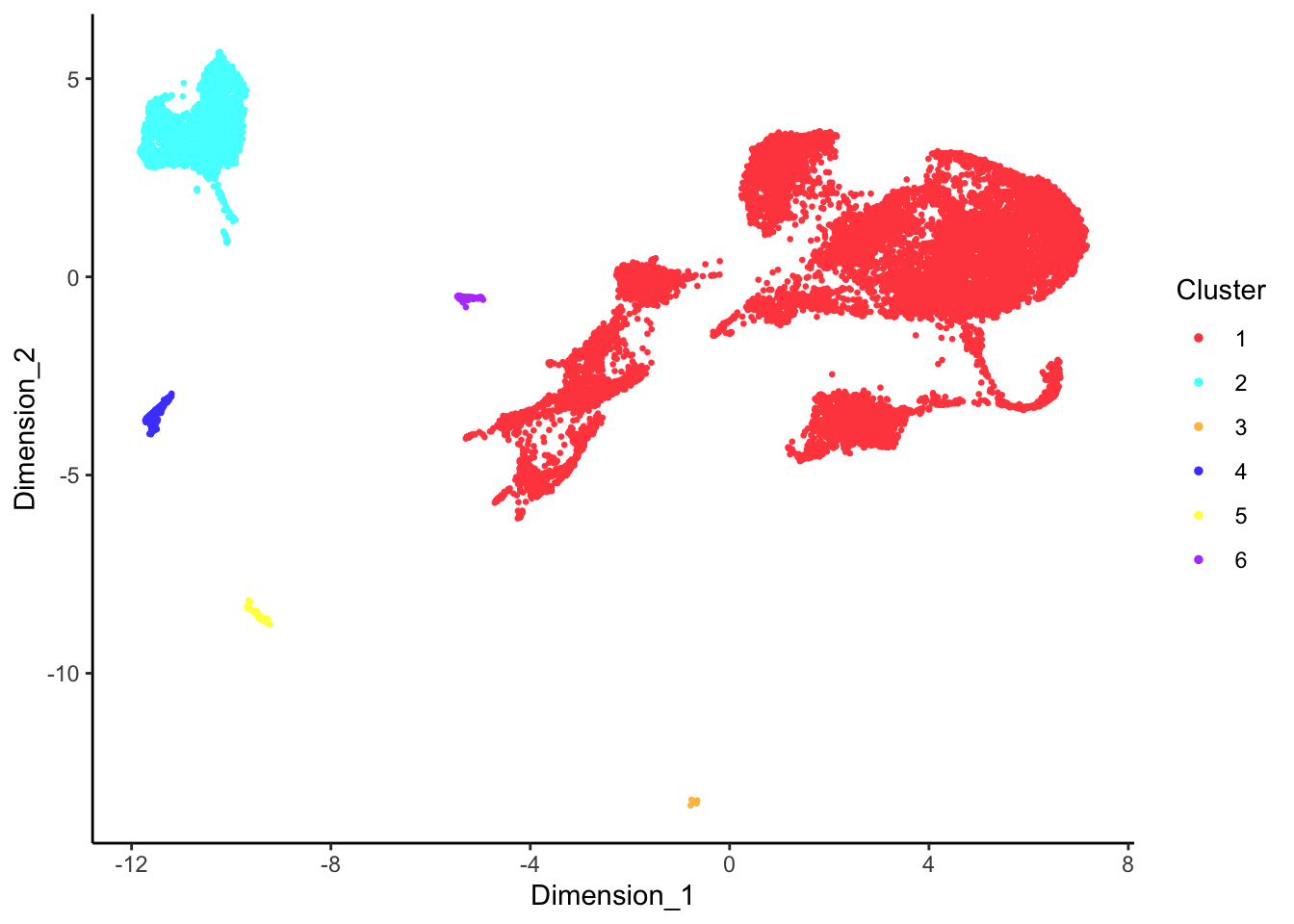
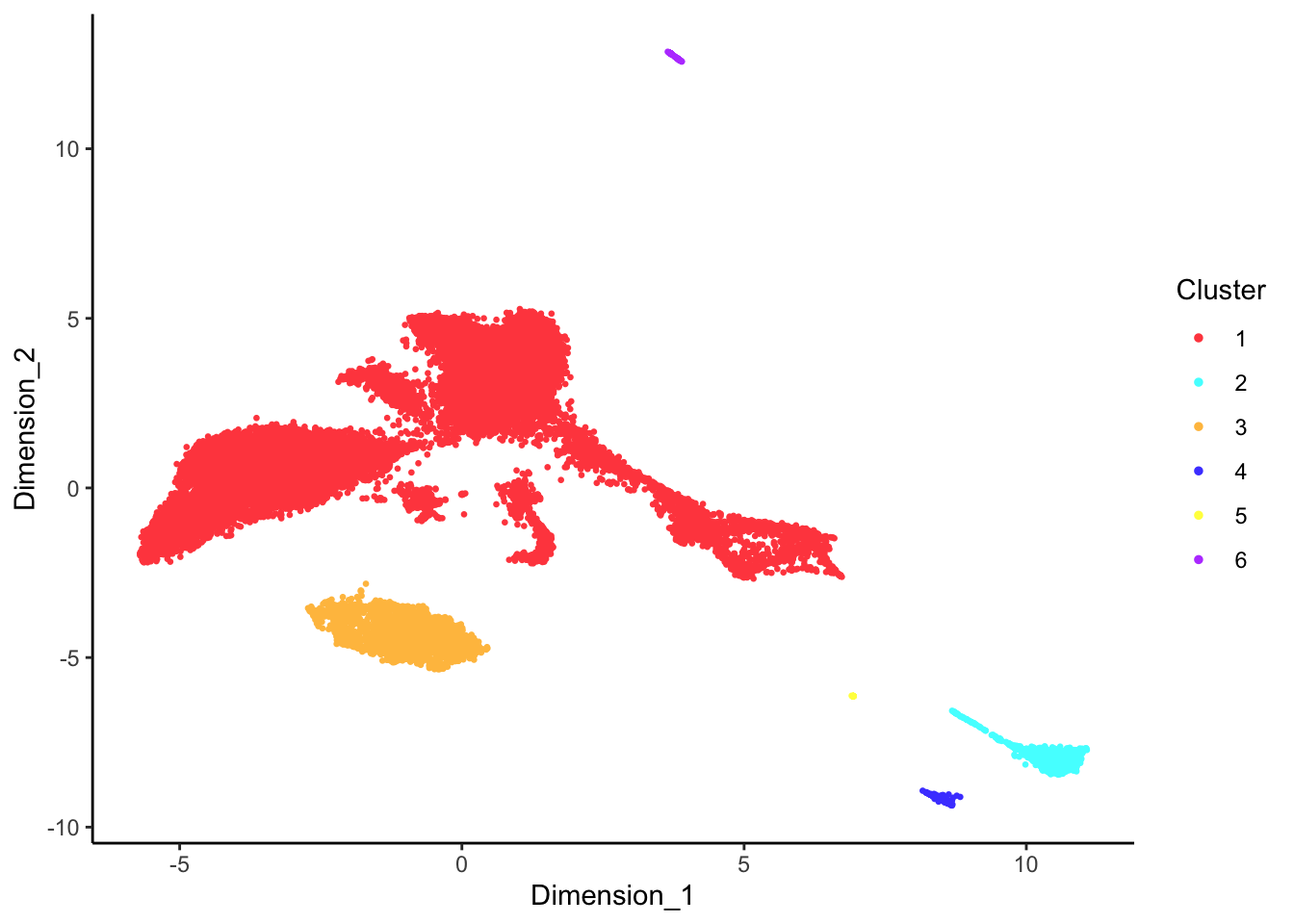
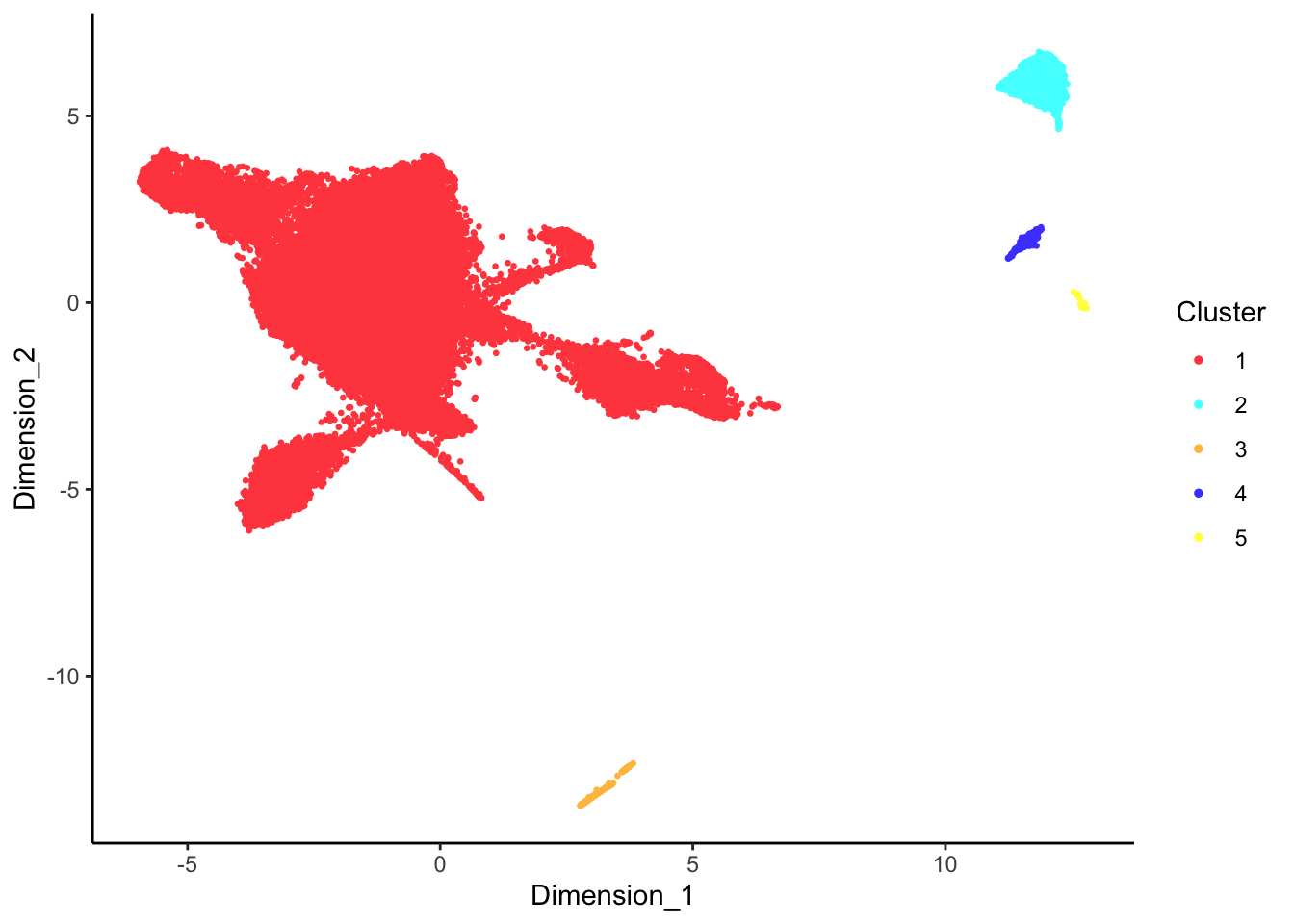
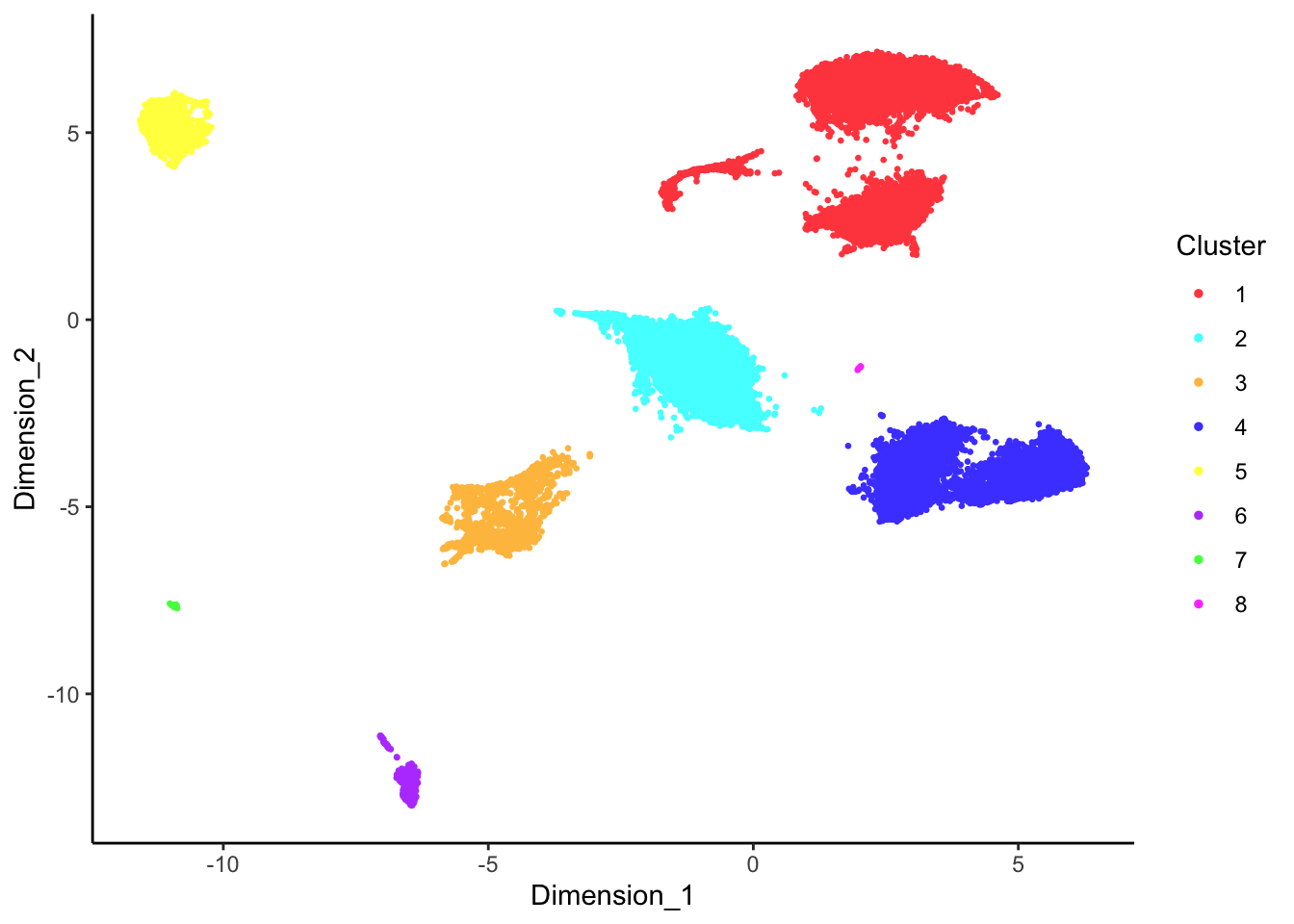
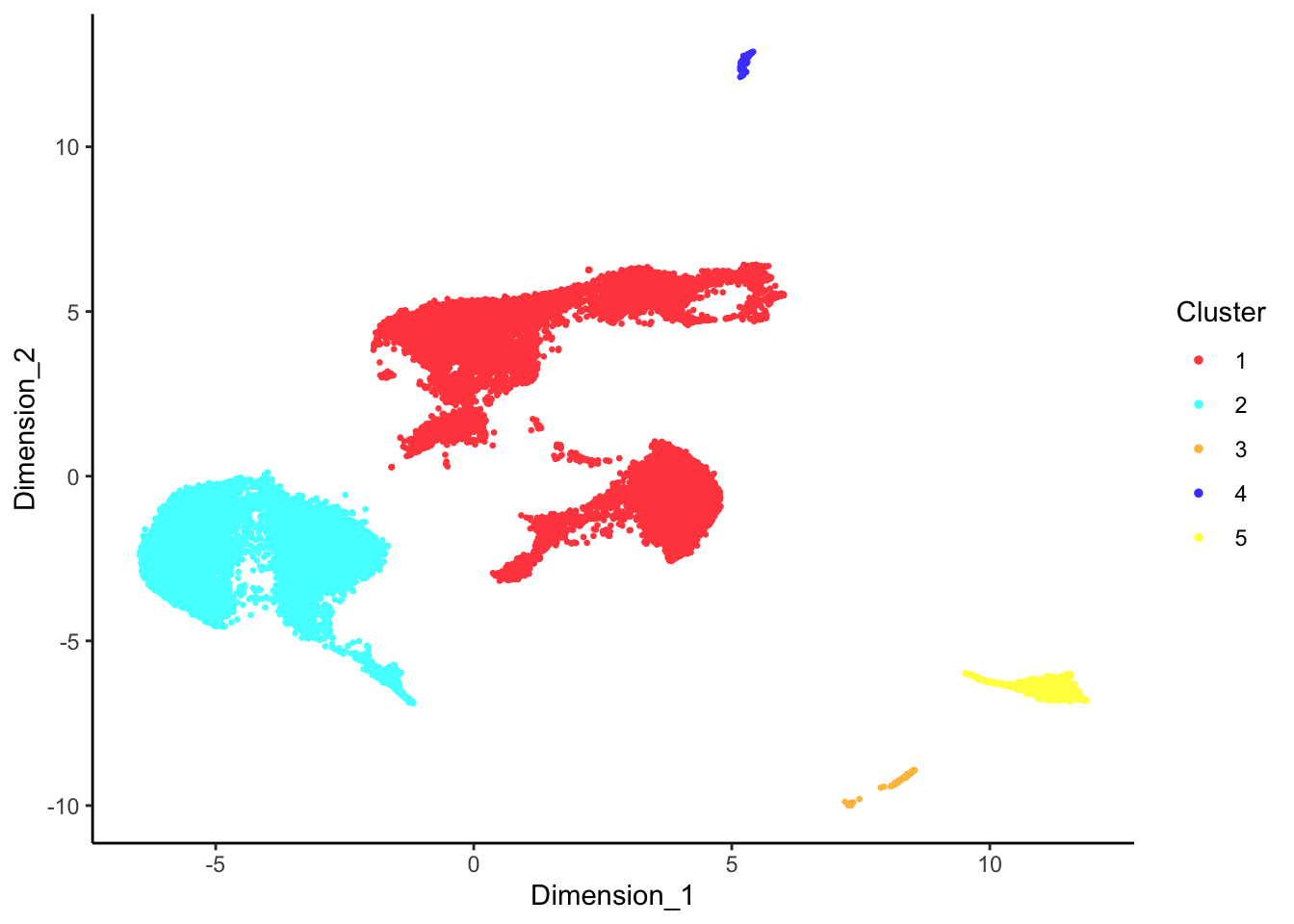
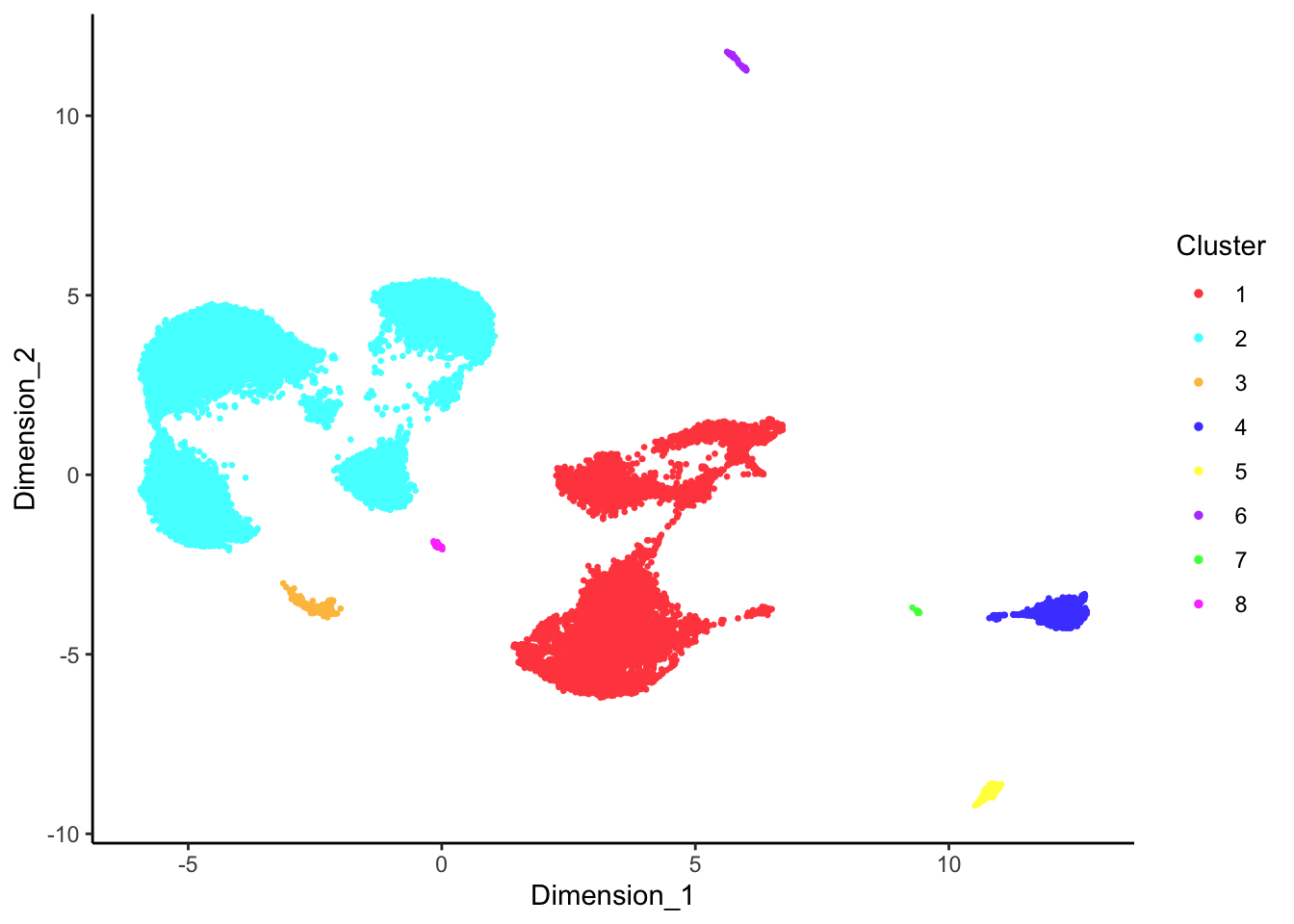
DecontX clusters
p <- lapply(1:length(sceLst), function(i){
sce <- sceLst[[i]]
if(length(levels(sce$Capture)) < 4){
umap <- reducedDim(sce, glue::glue("decontX_UMAP"))
plotDimReduceCluster(x = sce$decontX_clusters,
dim1 = umap[, 1], dim2 = umap[, 2])
} else {
capture_names <- levels(sce$Capture)
p <- lapply(capture_names, function(cn){
umap <- reducedDim(sce, glue::glue("decontX_{cn}_UMAP"))
plotDimReduceCluster(x = sce$decontX_clusters,
dim1 = umap[, 1], dim2 = umap[, 2])
})
wrap_plots(p, ncol = 2)
}
})
p[[1]]
Past versions of unnamed-chunk-4-1.png
Version
Author
Date
8abff7b
Jovana Maksimovic
2024-02-28
[[2]]
Past versions of unnamed-chunk-4-2.png
Version
Author
Date
8abff7b
Jovana Maksimovic
2024-02-28
[[3]]
Past versions of unnamed-chunk-4-3.png
Version
Author
Date
8abff7b
Jovana Maksimovic
2024-02-28
[[4]]
Past versions of unnamed-chunk-4-4.png
Version
Author
Date
8abff7b
Jovana Maksimovic
2024-02-28
[[5]]
Past versions of unnamed-chunk-4-5.png
Version
Author
Date
8abff7b
Jovana Maksimovic
2024-02-28
[[6]]
Past versions of unnamed-chunk-4-6.png
Version
Author
Date
8abff7b
Jovana Maksimovic
2024-02-28
[[7]]
Past versions of unnamed-chunk-4-7.png
Version
Author
Date
8abff7b
Jovana Maksimovic
2024-02-28
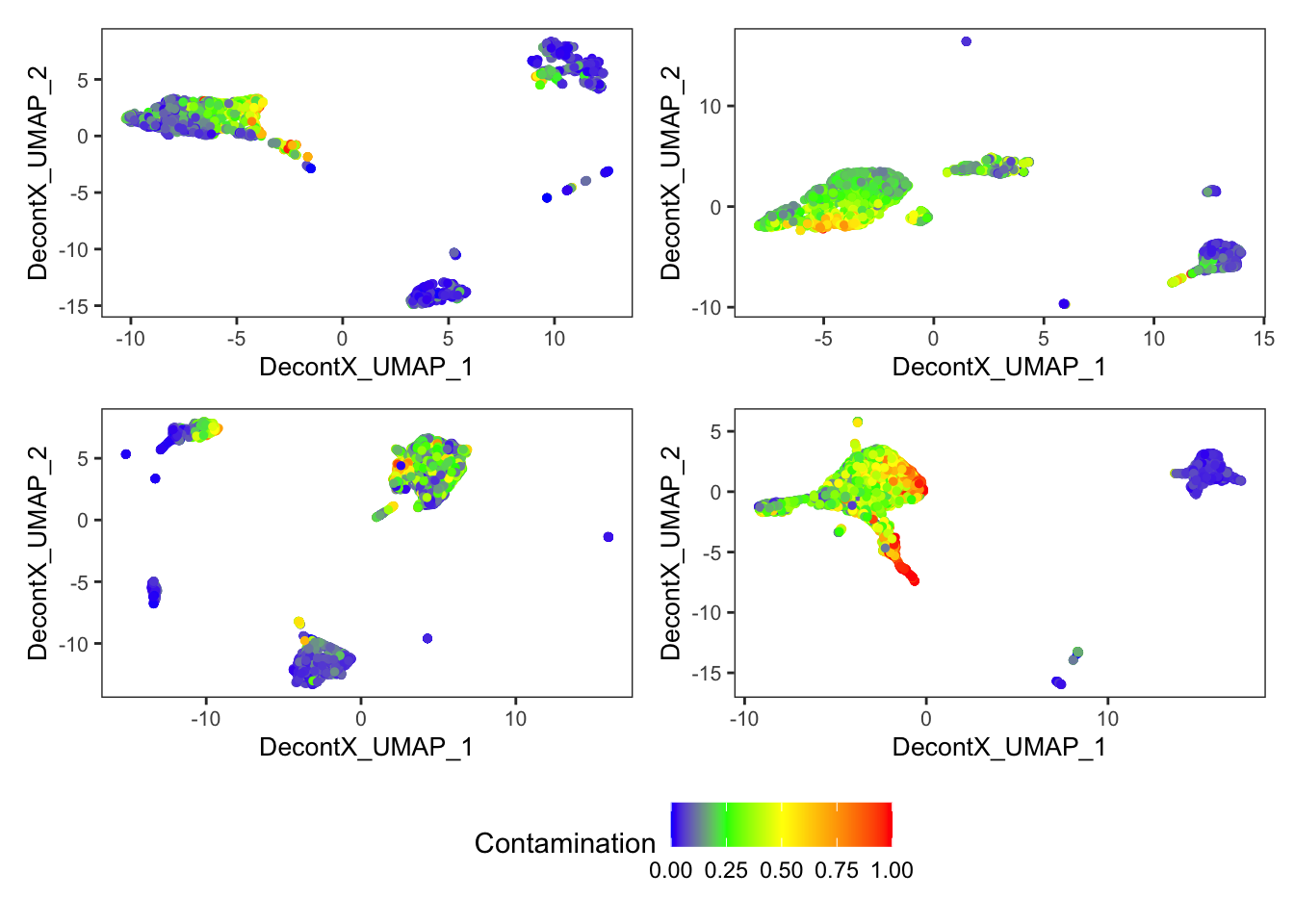
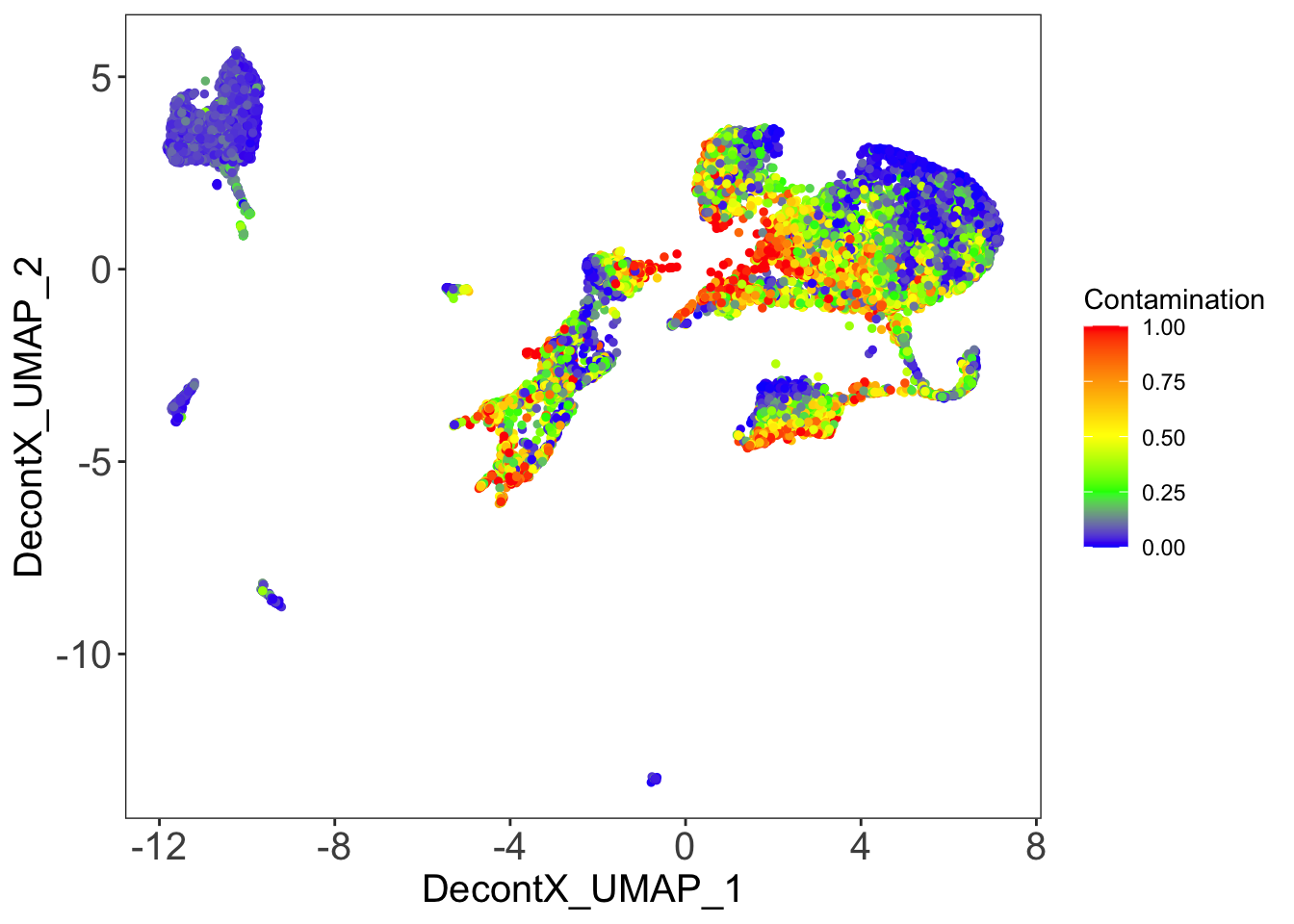
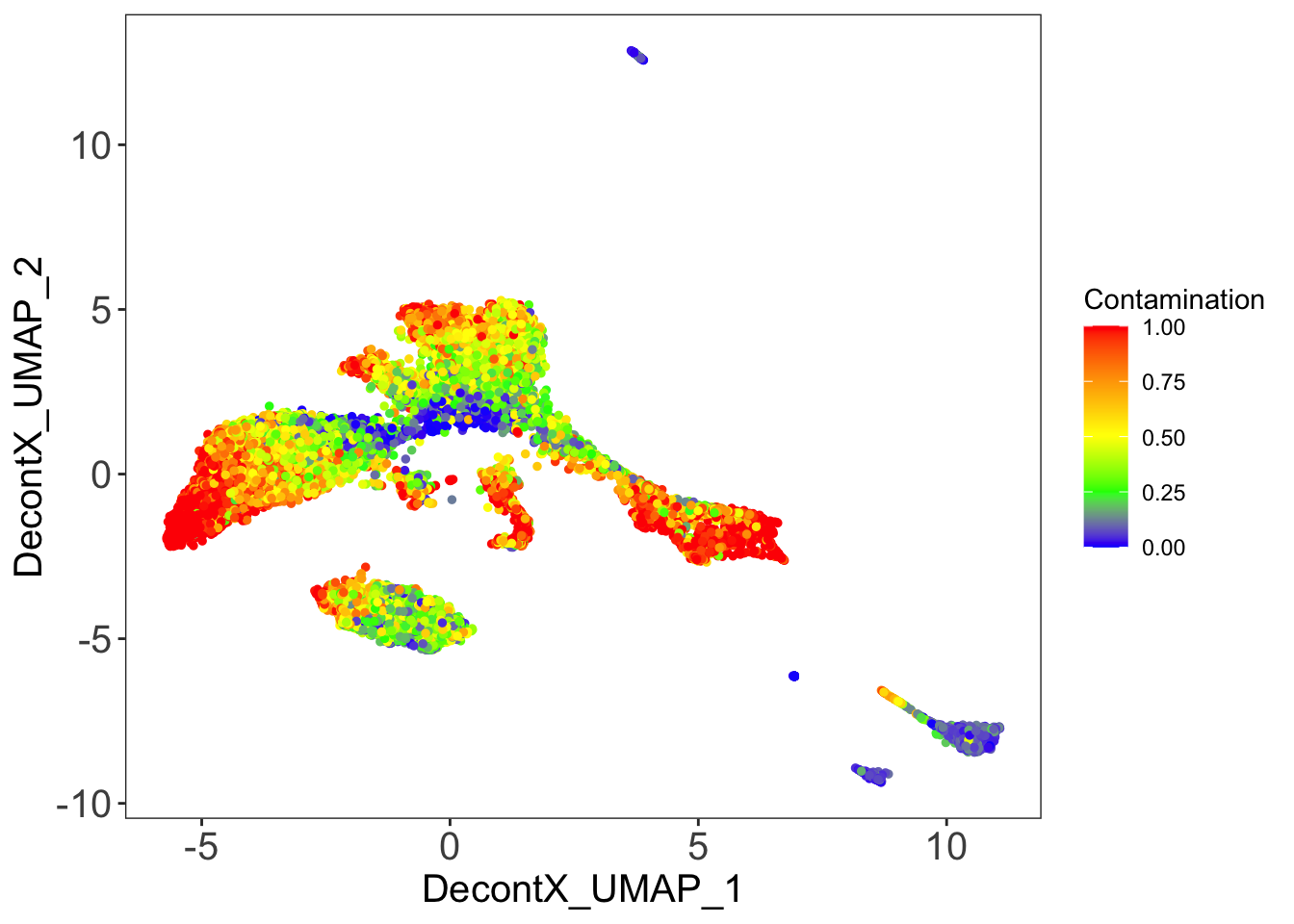
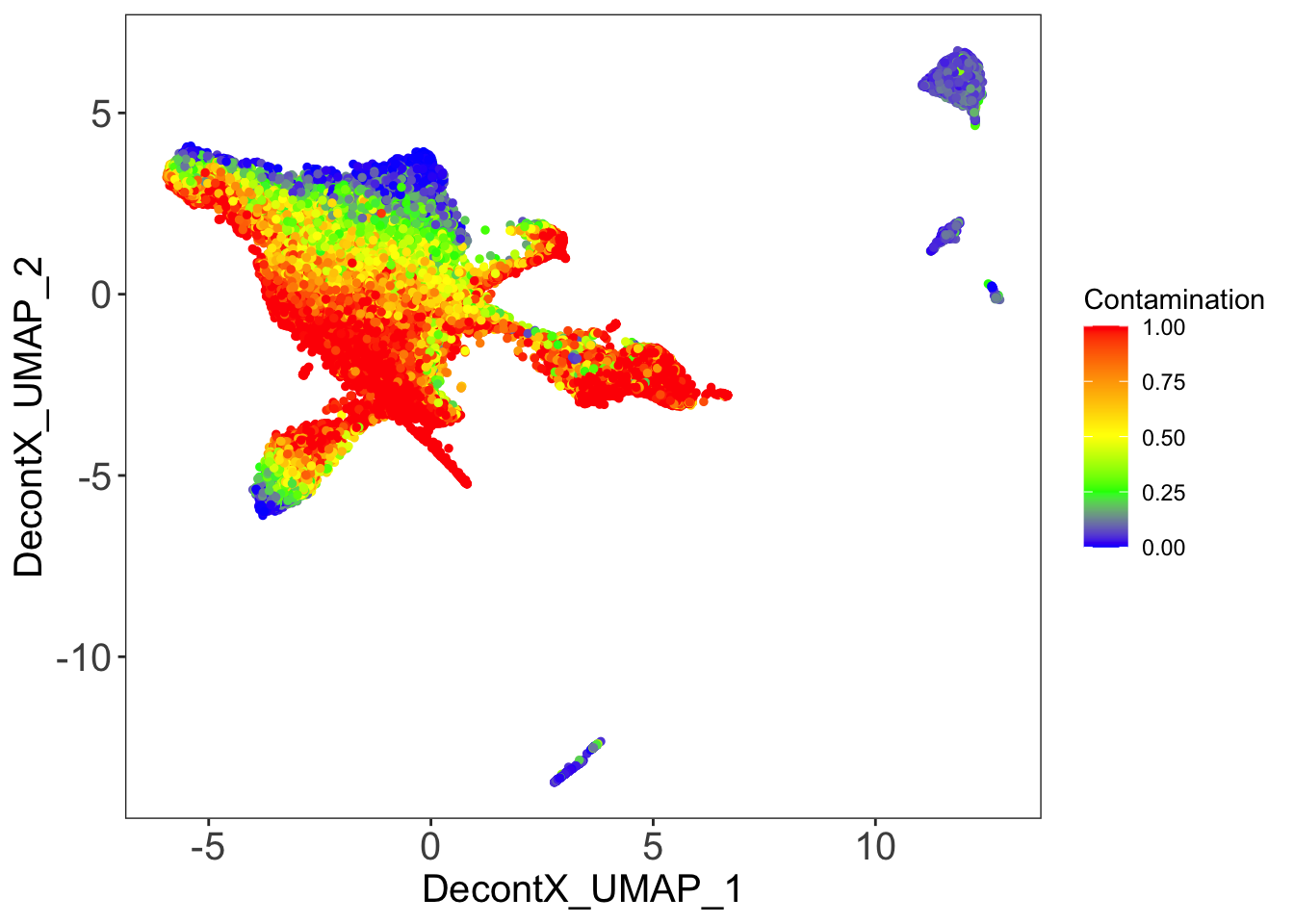
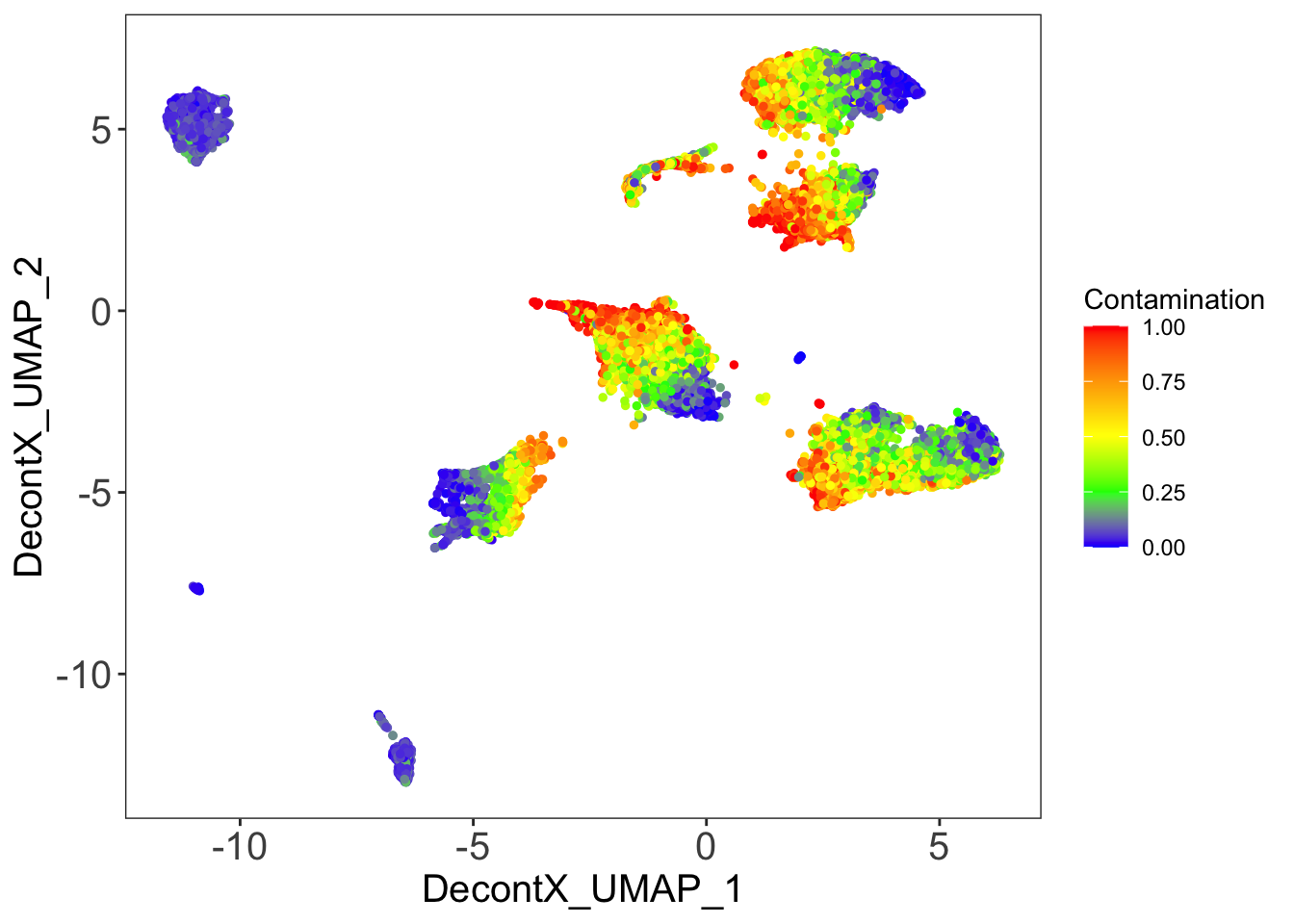
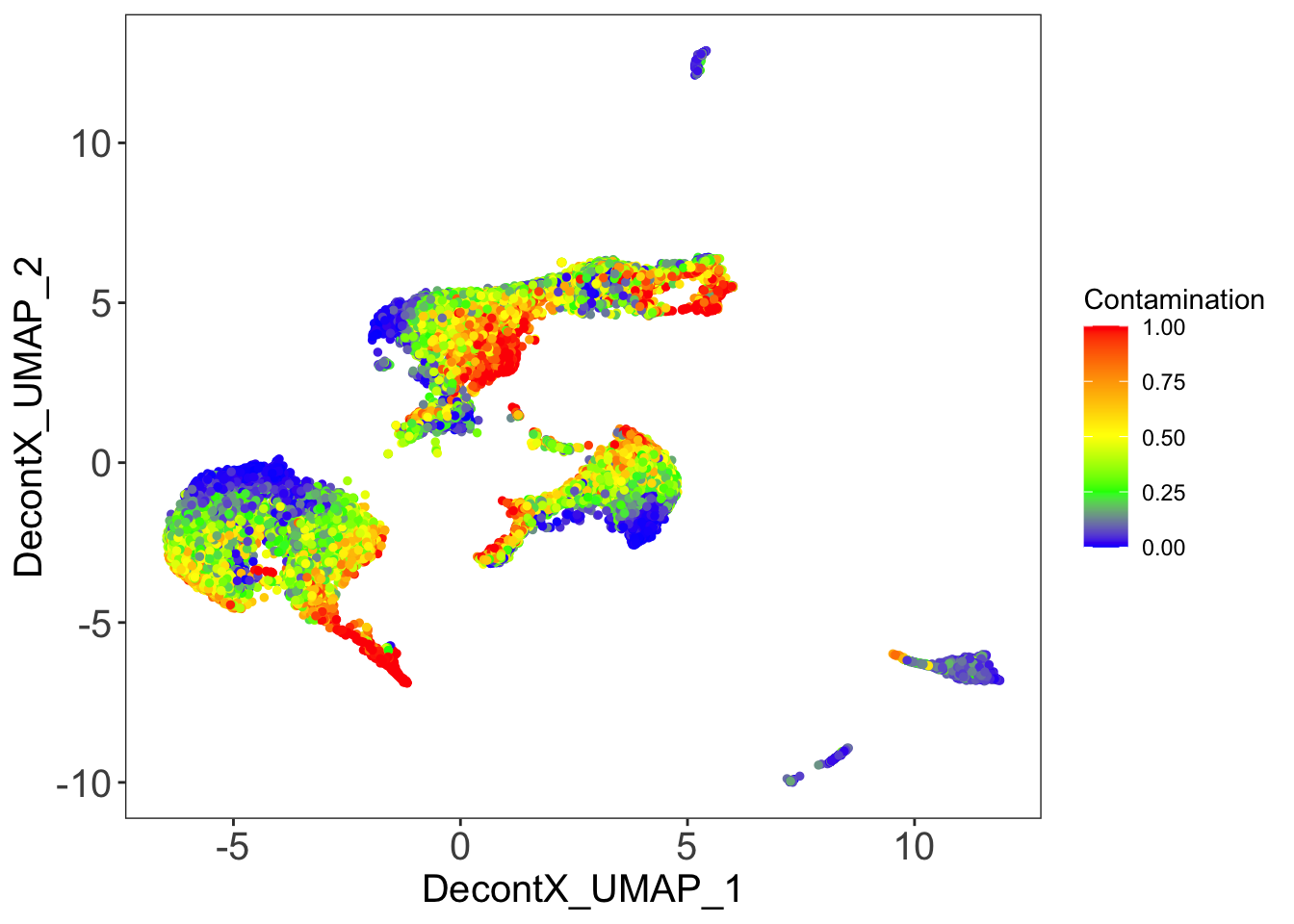
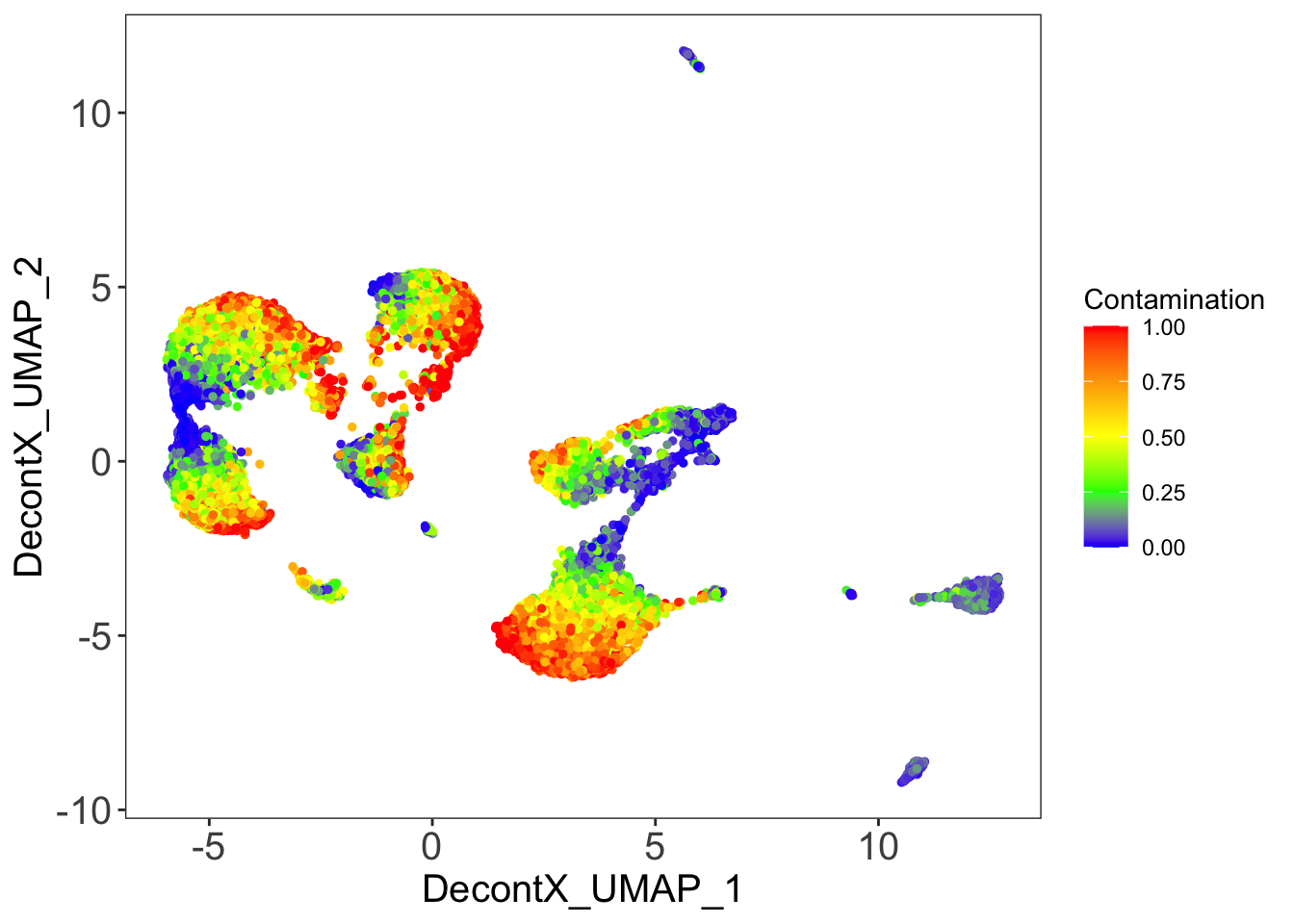
DecontX contamination
p <- lapply(1:length(sceLst), function(i){
sce <- sceLst[[i]]
if(length(levels(sce$Capture)) < 4){
plotDecontXContamination(sce)
} else {
capture_names <- levels(sce$Capture)
p <- lapply(capture_names, function(cn){
plotDecontXContamination(sce, batch = cn)
})
wrap_plots(p, ncol = 2, guides = "collect") &
theme(legend.position = "bottom",
axis.title = element_text(size = 10),
axis.text = element_text(size = 8))
}
})
p[[1]]
Past versions of unnamed-chunk-5-1.png
Version
Author
Date
8abff7b
Jovana Maksimovic
2024-02-28
[[2]]
Past versions of unnamed-chunk-5-2.png
Version
Author
Date
8abff7b
Jovana Maksimovic
2024-02-28
[[3]]
Past versions of unnamed-chunk-5-3.png
Version
Author
Date
8abff7b
Jovana Maksimovic
2024-02-28
[[4]]
Past versions of unnamed-chunk-5-4.png
Version
Author
Date
8abff7b
Jovana Maksimovic
2024-02-28
[[5]]
Past versions of unnamed-chunk-5-5.png
Version
Author
Date
8abff7b
Jovana Maksimovic
2024-02-28
[[6]]
Past versions of unnamed-chunk-5-6.png
Version
Author
Date
8abff7b
Jovana Maksimovic
2024-02-28
[[7]]
Past versions of unnamed-chunk-5-7.png
Version
Author
Date
8abff7b
Jovana Maksimovic
2024-02-28
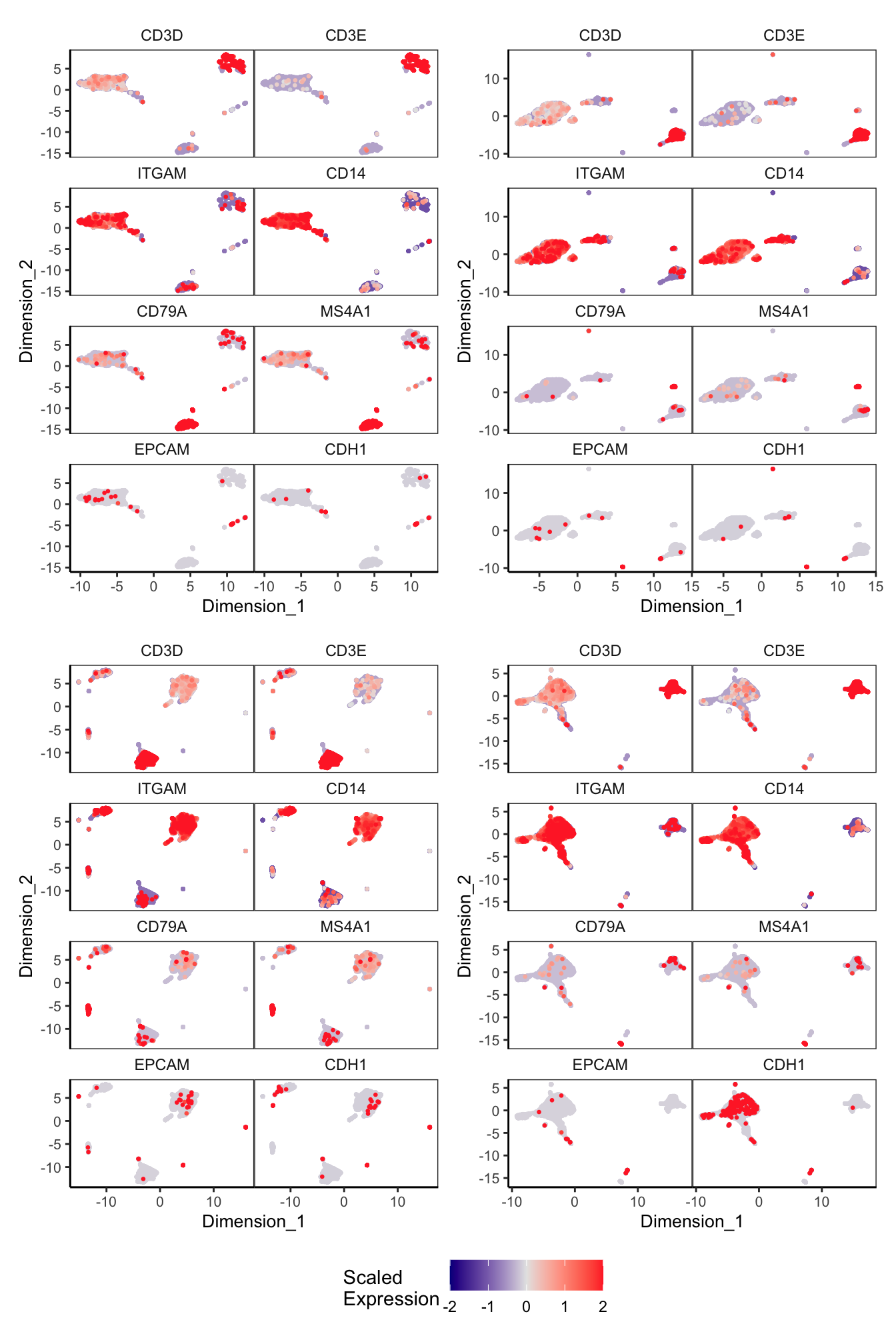
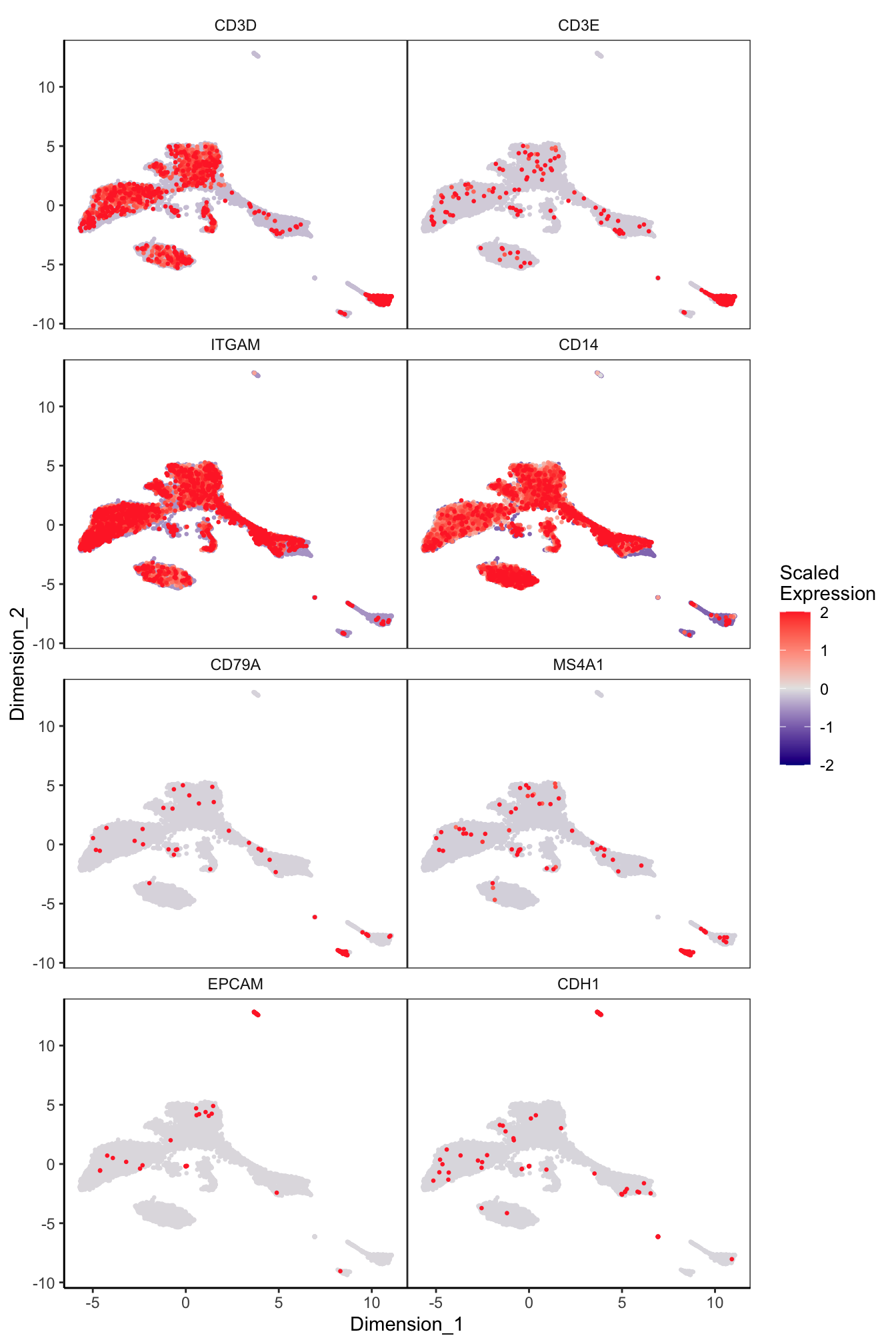
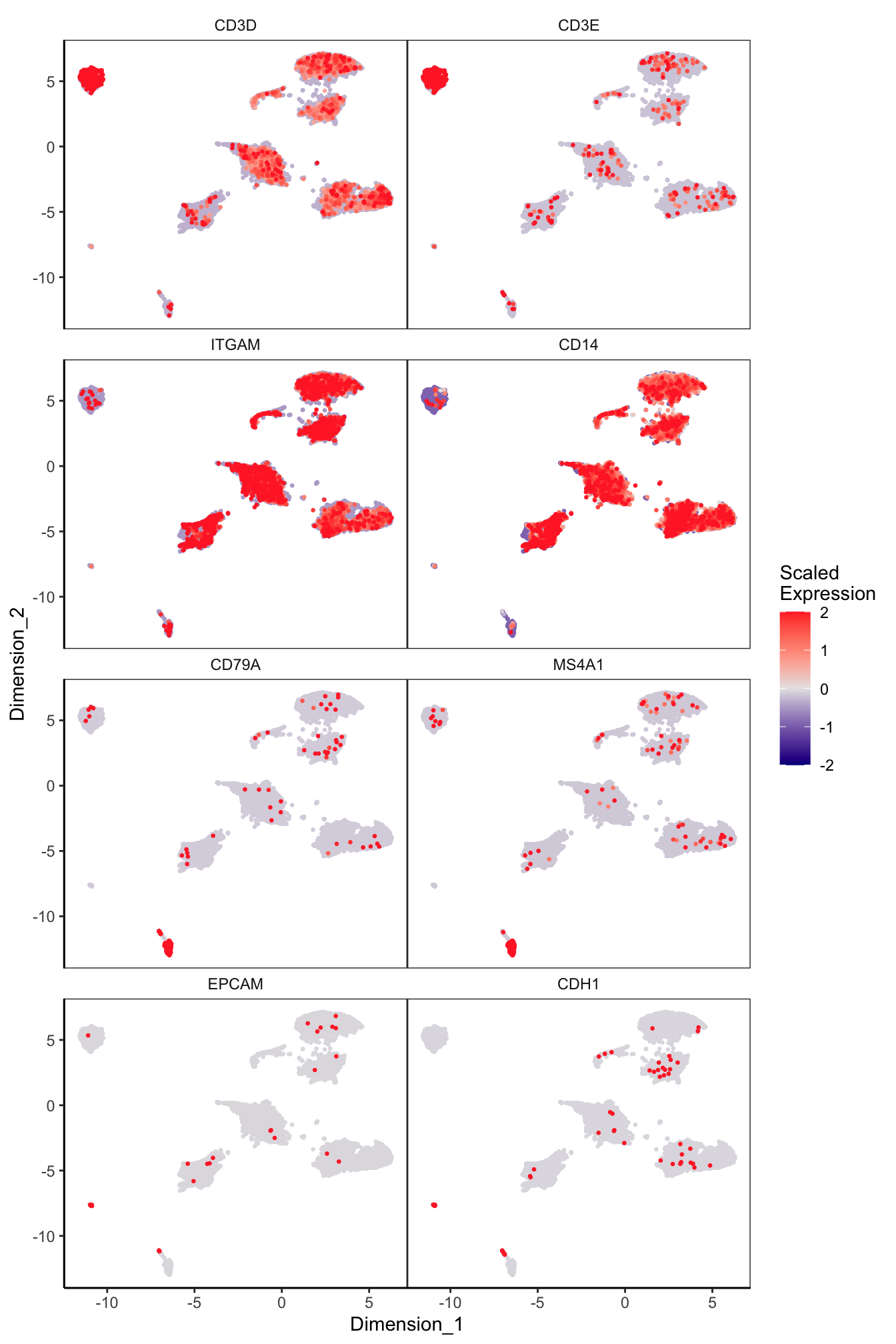
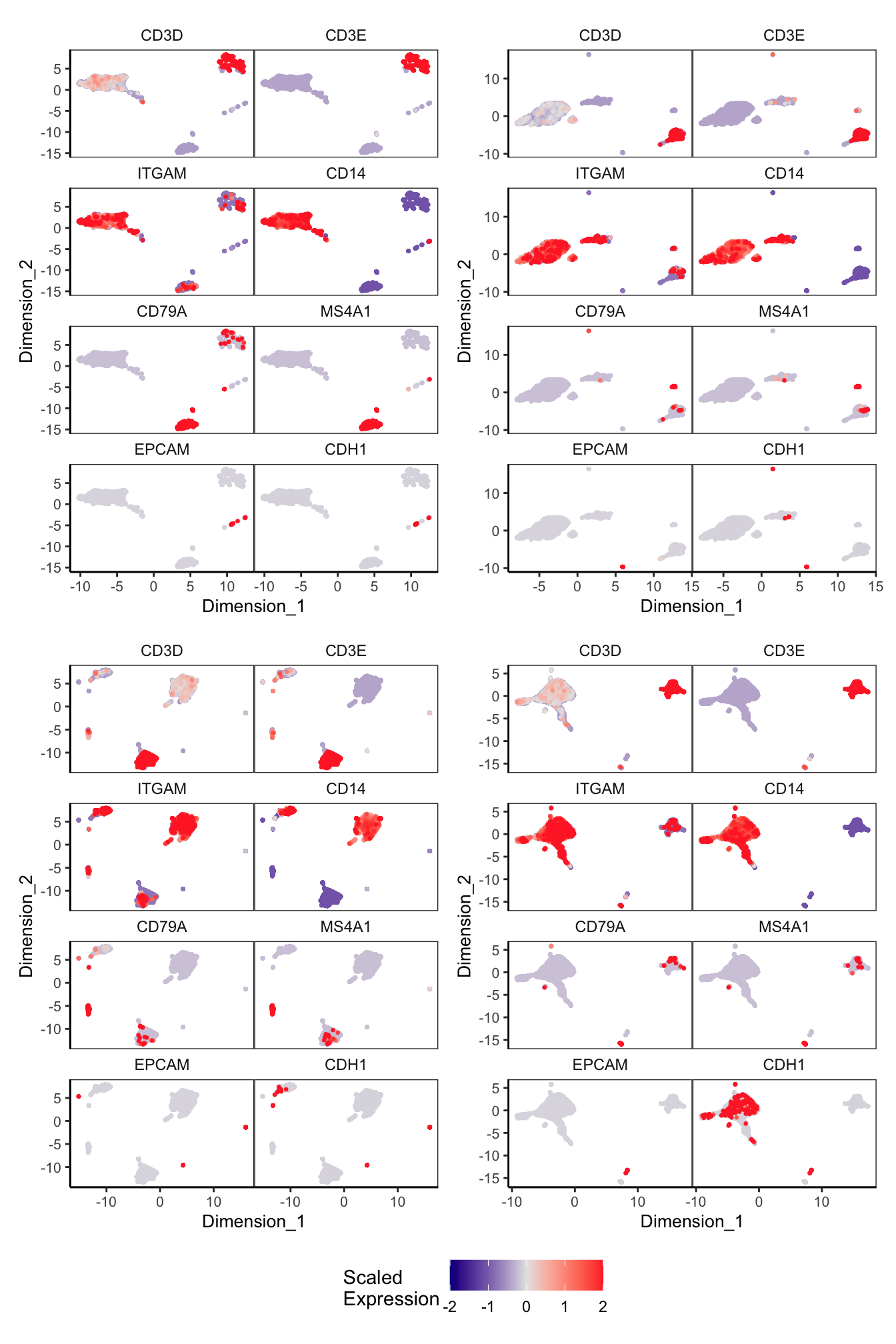
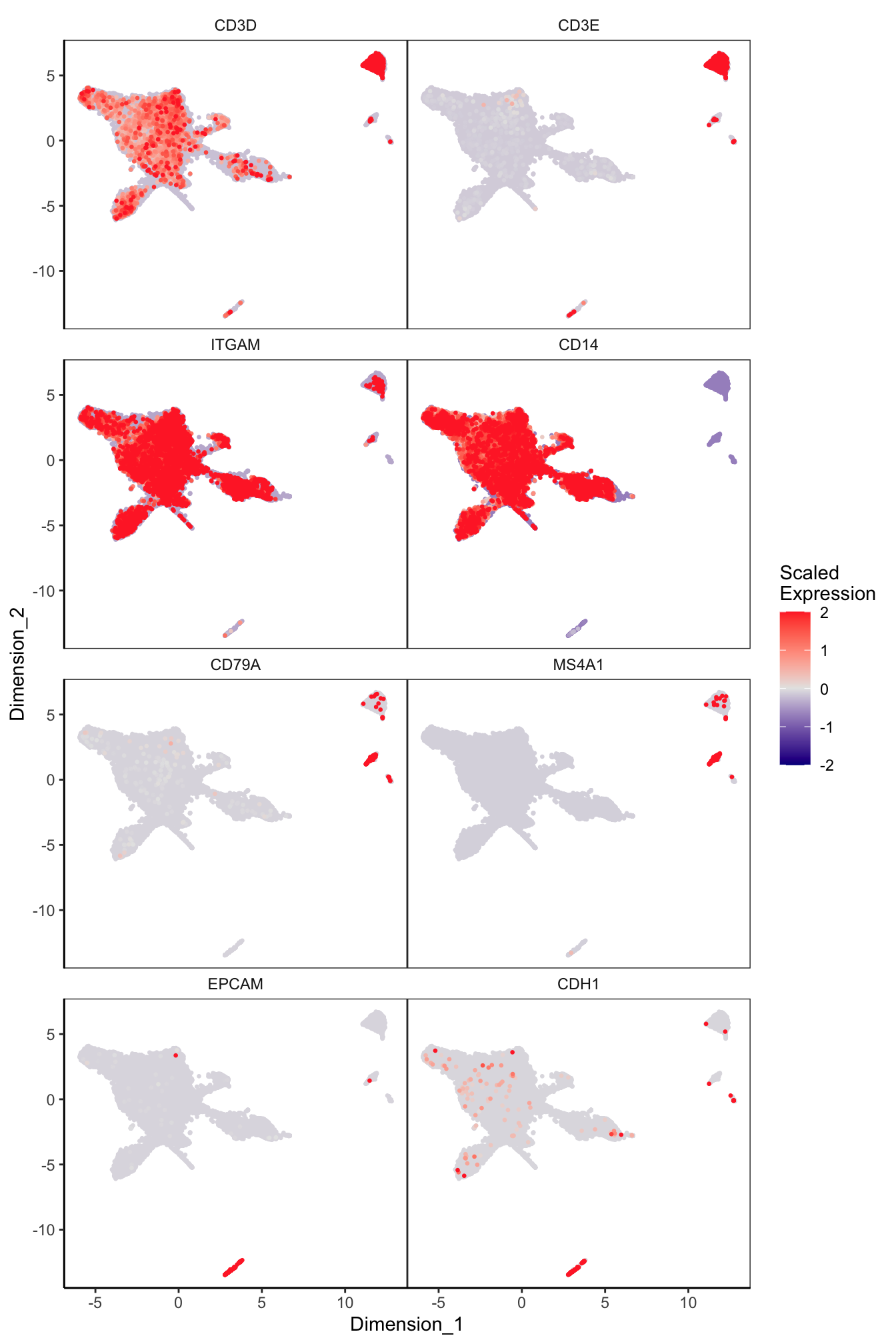
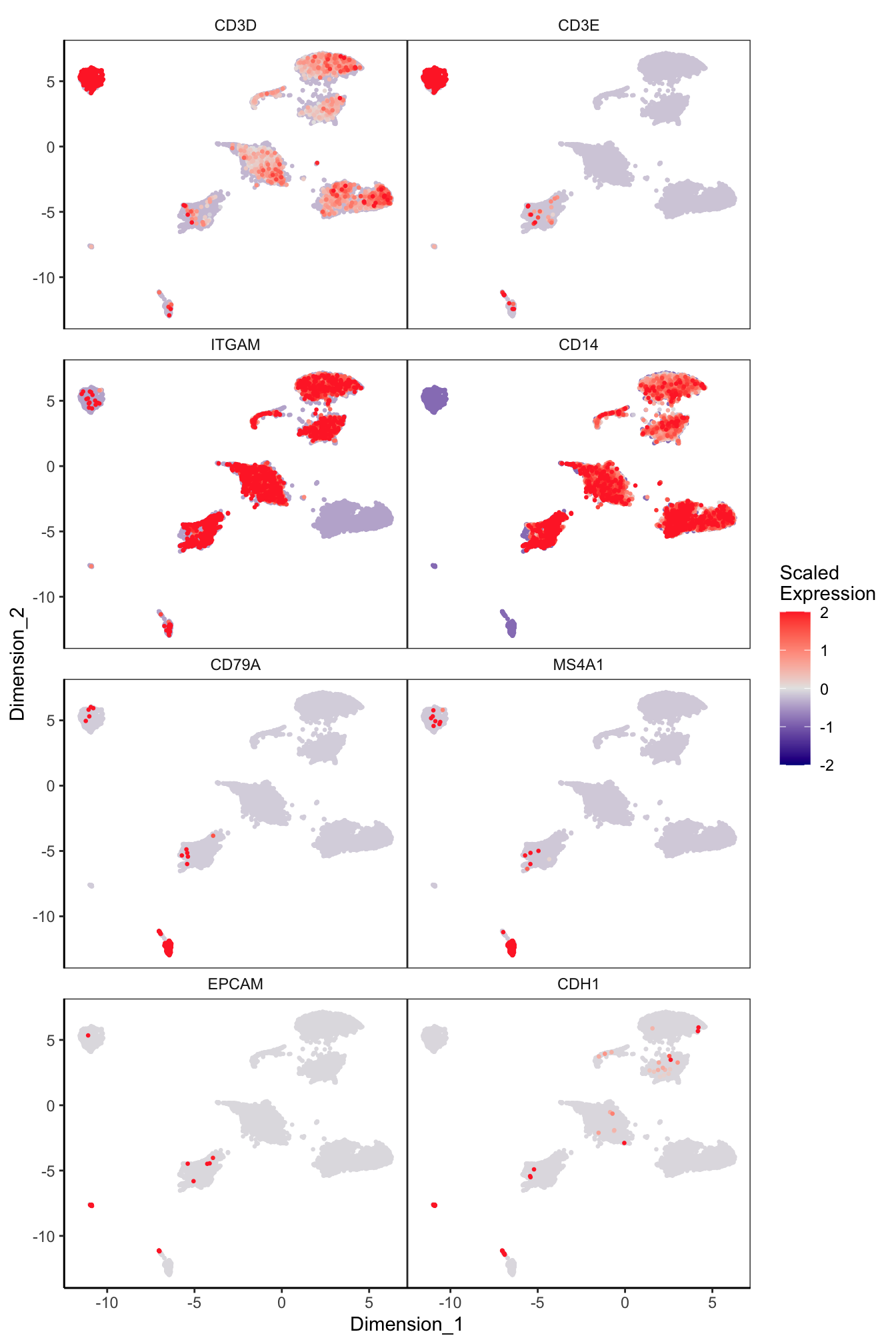
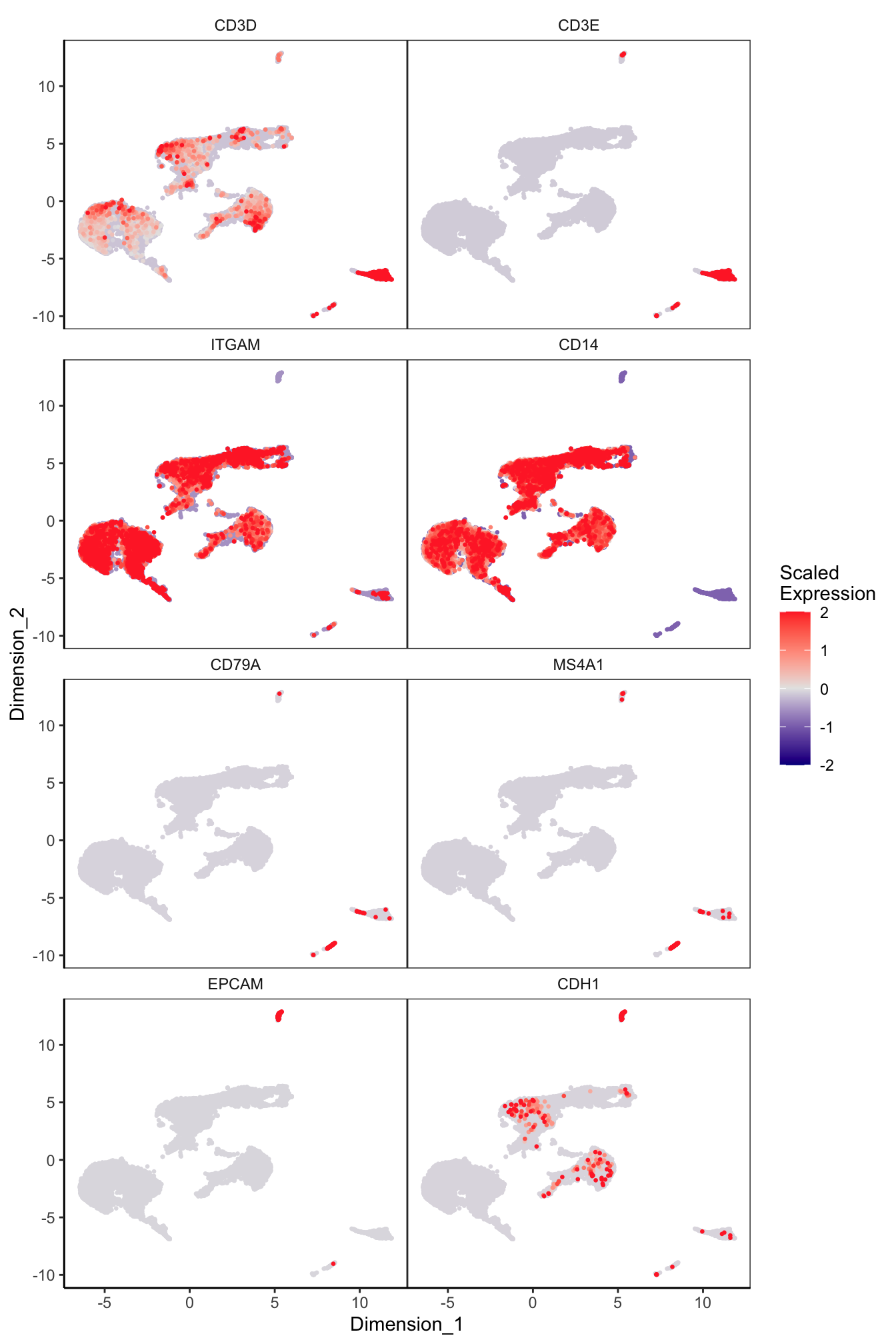
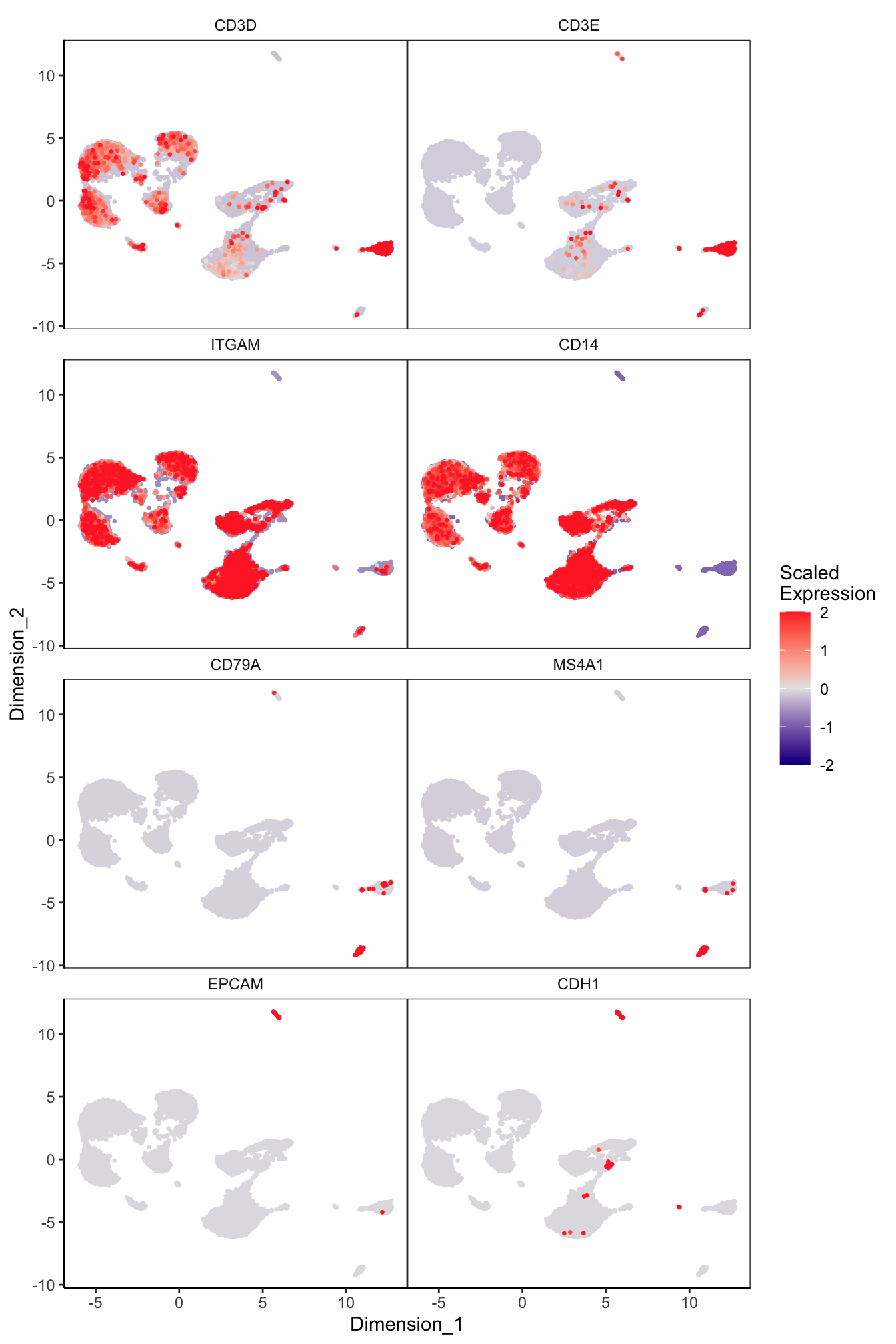
Main cell type markers (before decontX)
p <- lapply(1:length(sceLst), function(i){
sce <- sceLst[[i]]
if(length(levels(sce$Capture)) < 4){
sce_decont <- logNormCounts(sce)
rownames(sce_decont) <- rowData(sce_decont)$Symbol
umap <- reducedDim(sce_decont, glue::glue("decontX_UMAP"))
plotDimReduceFeature(as.matrix(logcounts(sce_decont)),
dim1 = umap[, 1],
dim2 = umap[, 2],
features = c("CD3D", "CD3E", # T-cells
"ITGAM", "CD14", # Macs
"CD79A", "MS4A1", # B-cells
"EPCAM", "CDH1"), # Epithelial
exactMatch = TRUE,
ncol = 2)
} else {
sce_decont <- logNormCounts(sce)
rownames(sce_decont) <- rowData(sce_decont)$Symbol
capture_names <- levels(sce$Capture)
p <- lapply(capture_names, function(cn){
umap <- reducedDim(sce_decont, glue::glue("decontX_{cn}_UMAP"))
plotDimReduceFeature(as.matrix(logcounts(sce_decont)),
dim1 = umap[, 1],
dim2 = umap[, 2],
features = c("CD3D", "CD3E", # T-cells
"ITGAM", "CD14", # Macs
"CD79A", "MS4A1", # B-cells
"EPCAM", "CDH1"), # Epithelial
exactMatch = TRUE,
ncol = 2)
})
wrap_plots(p, ncol = 2, guides = "collect") &
theme(legend.position = "bottom",
axis.title = element_text(size = 10),
axis.text = element_text(size = 8))
}
})
p[[1]]
Past versions of unnamed-chunk-6-1.png
Version
Author
Date
8abff7b
Jovana Maksimovic
2024-02-28
[[2]]
Past versions of unnamed-chunk-6-2.png
Version
Author
Date
8abff7b
Jovana Maksimovic
2024-02-28
[[3]]
Past versions of unnamed-chunk-6-3.png
Version
Author
Date
8abff7b
Jovana Maksimovic
2024-02-28
[[4]]
Past versions of unnamed-chunk-6-4.png
Version
Author
Date
8abff7b
Jovana Maksimovic
2024-02-28
[[5]]
Past versions of unnamed-chunk-6-5.png
Version
Author
Date
8abff7b
Jovana Maksimovic
2024-02-28
[[6]]
Past versions of unnamed-chunk-6-6.png
Version
Author
Date
8abff7b
Jovana Maksimovic
2024-02-28
[[7]]
Past versions of unnamed-chunk-6-7.png
Version
Author
Date
8abff7b
Jovana Maksimovic
2024-02-28
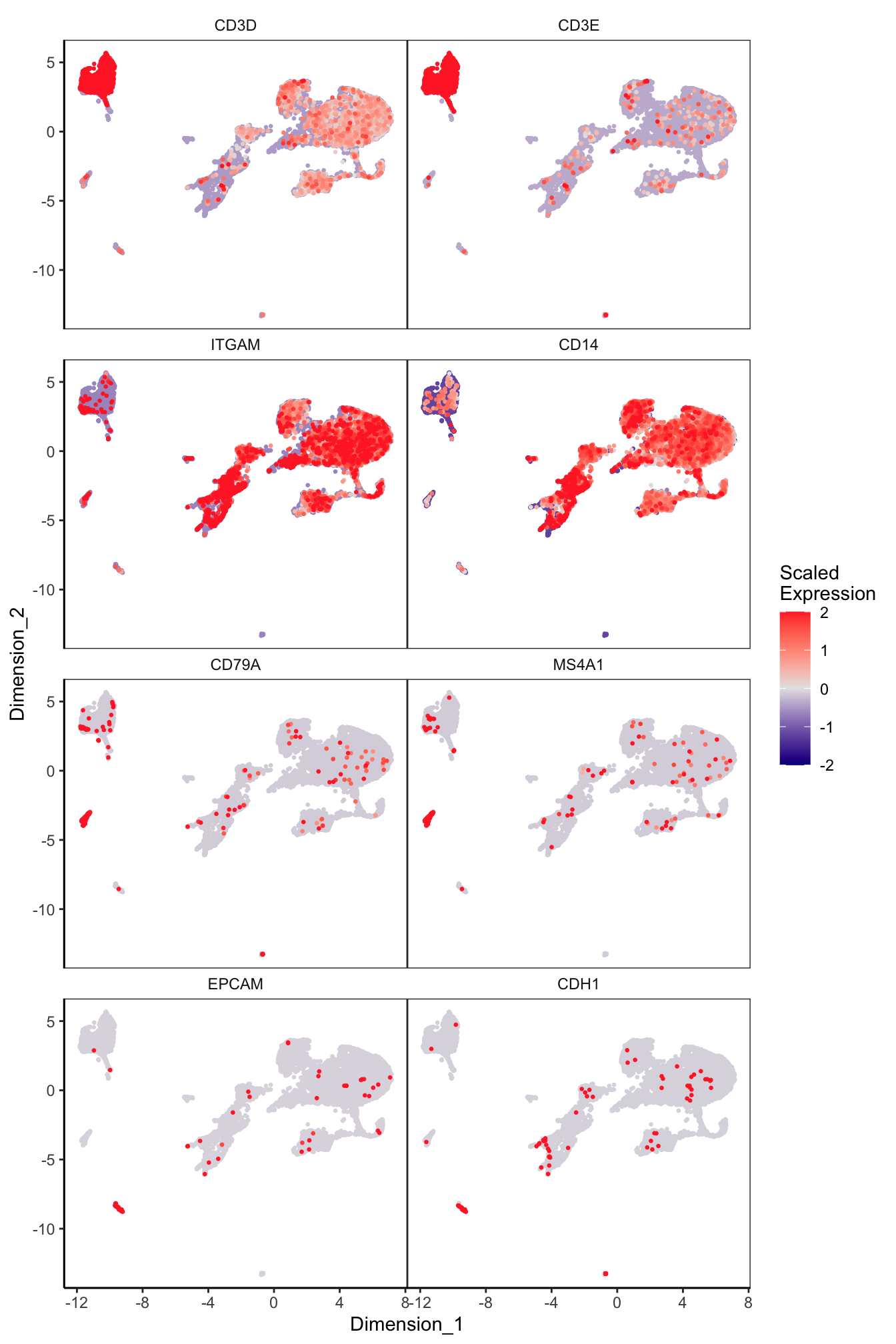
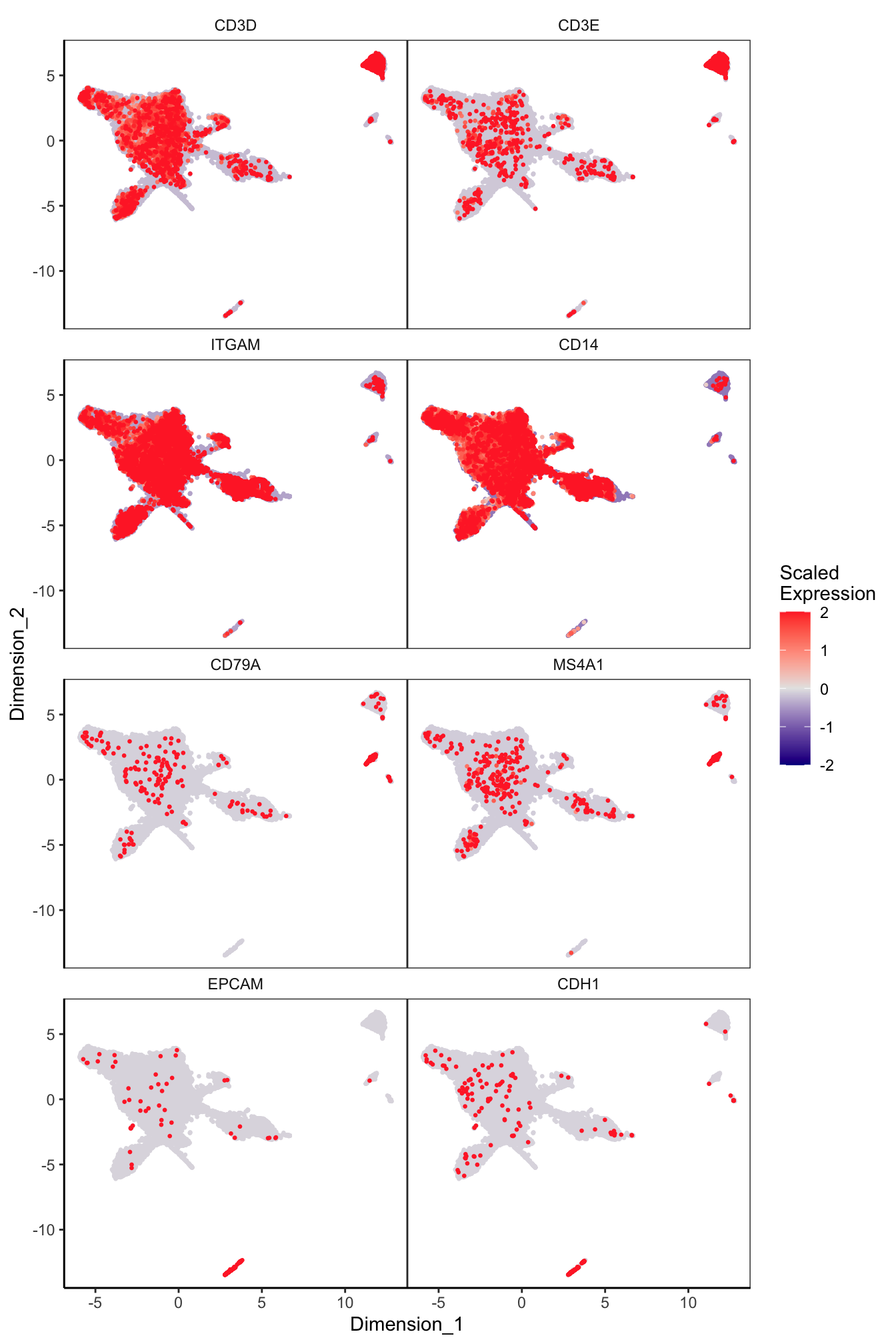
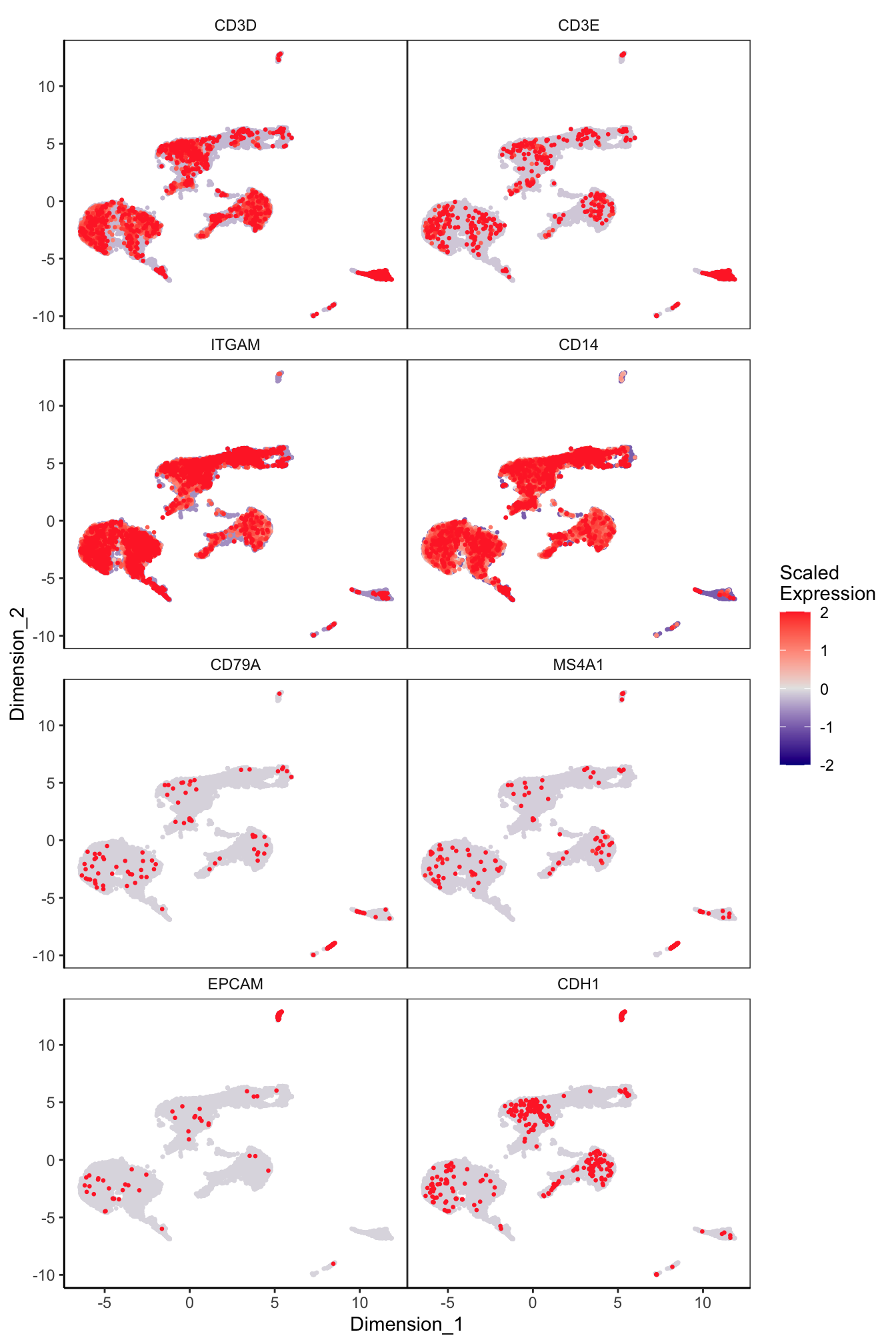
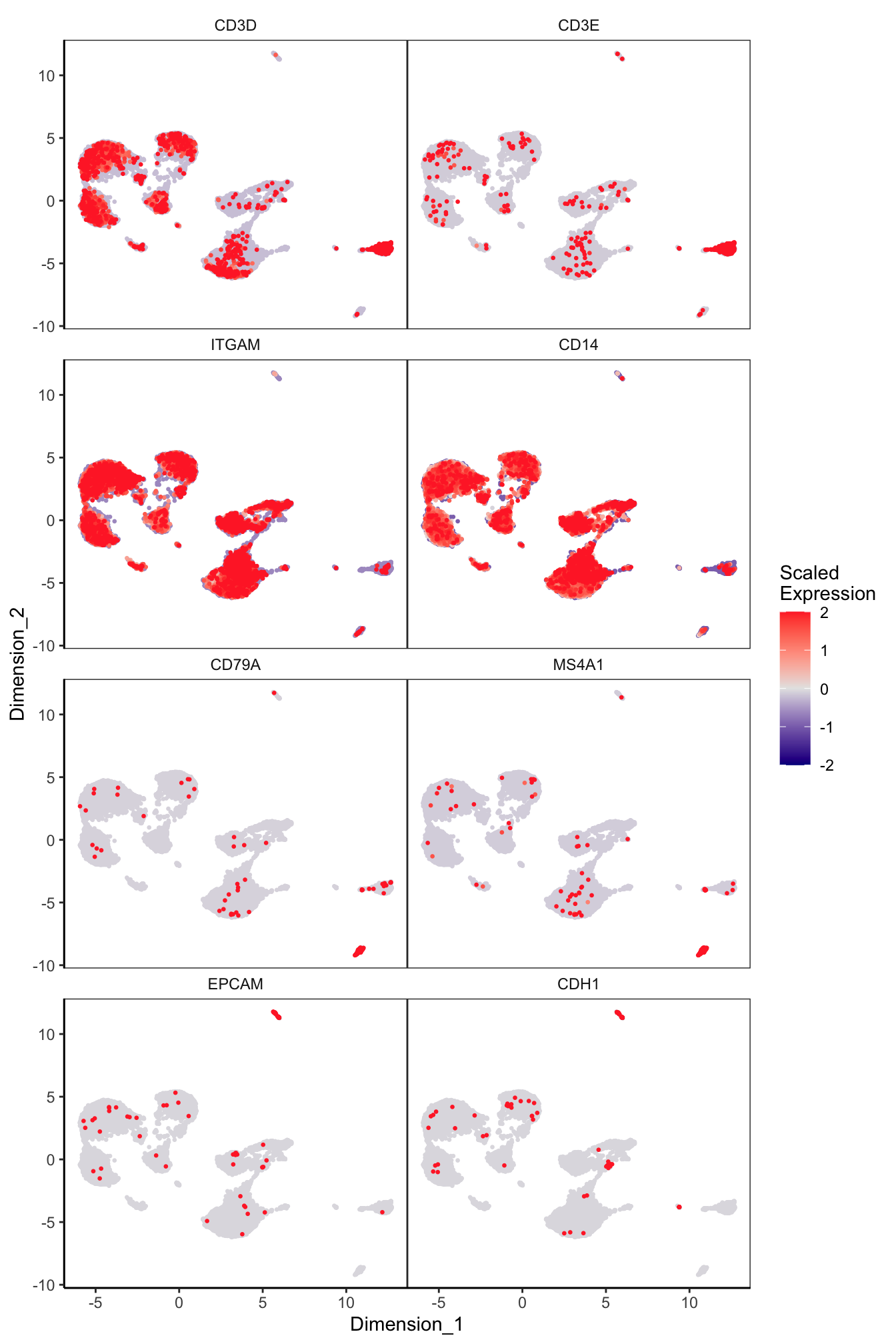
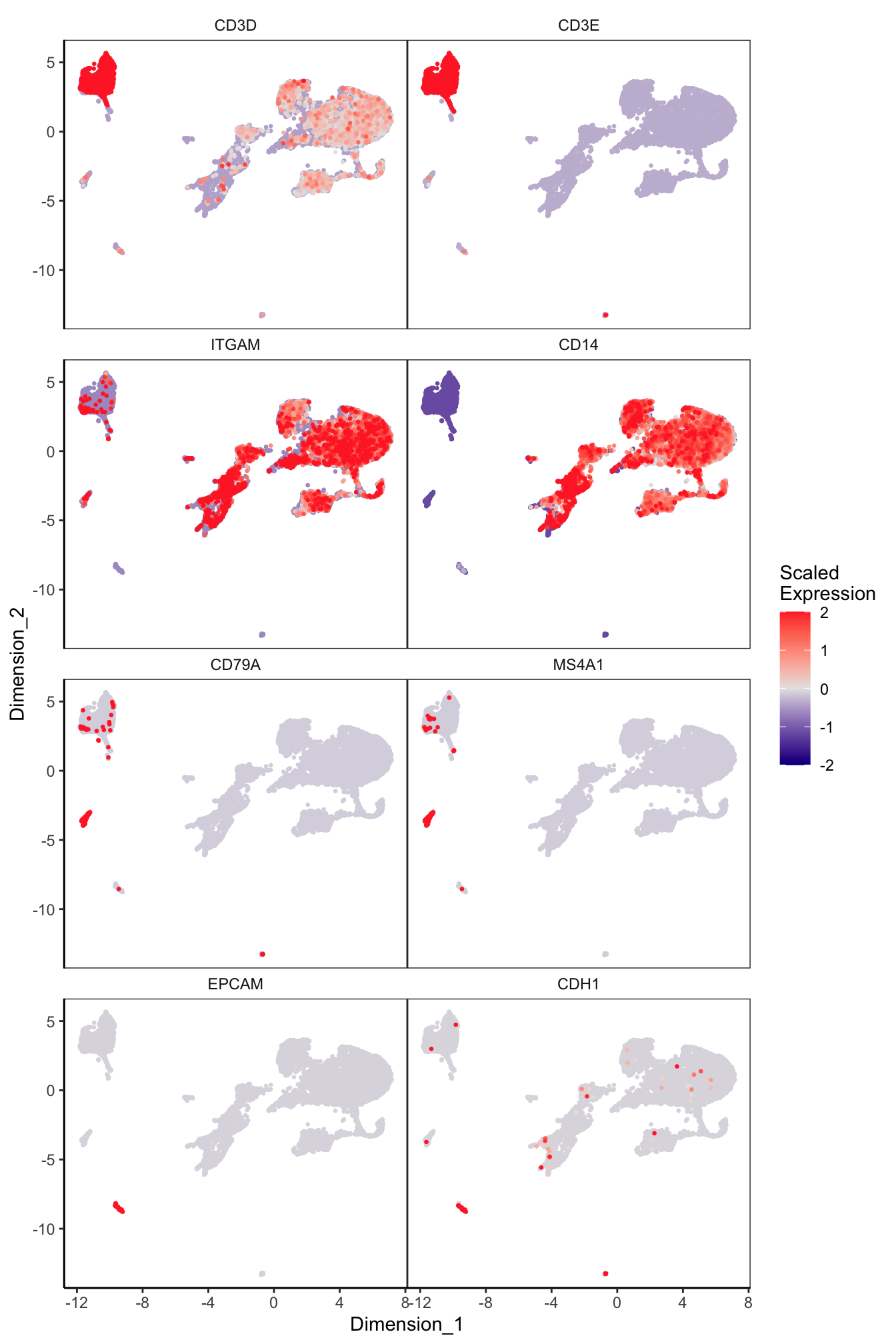
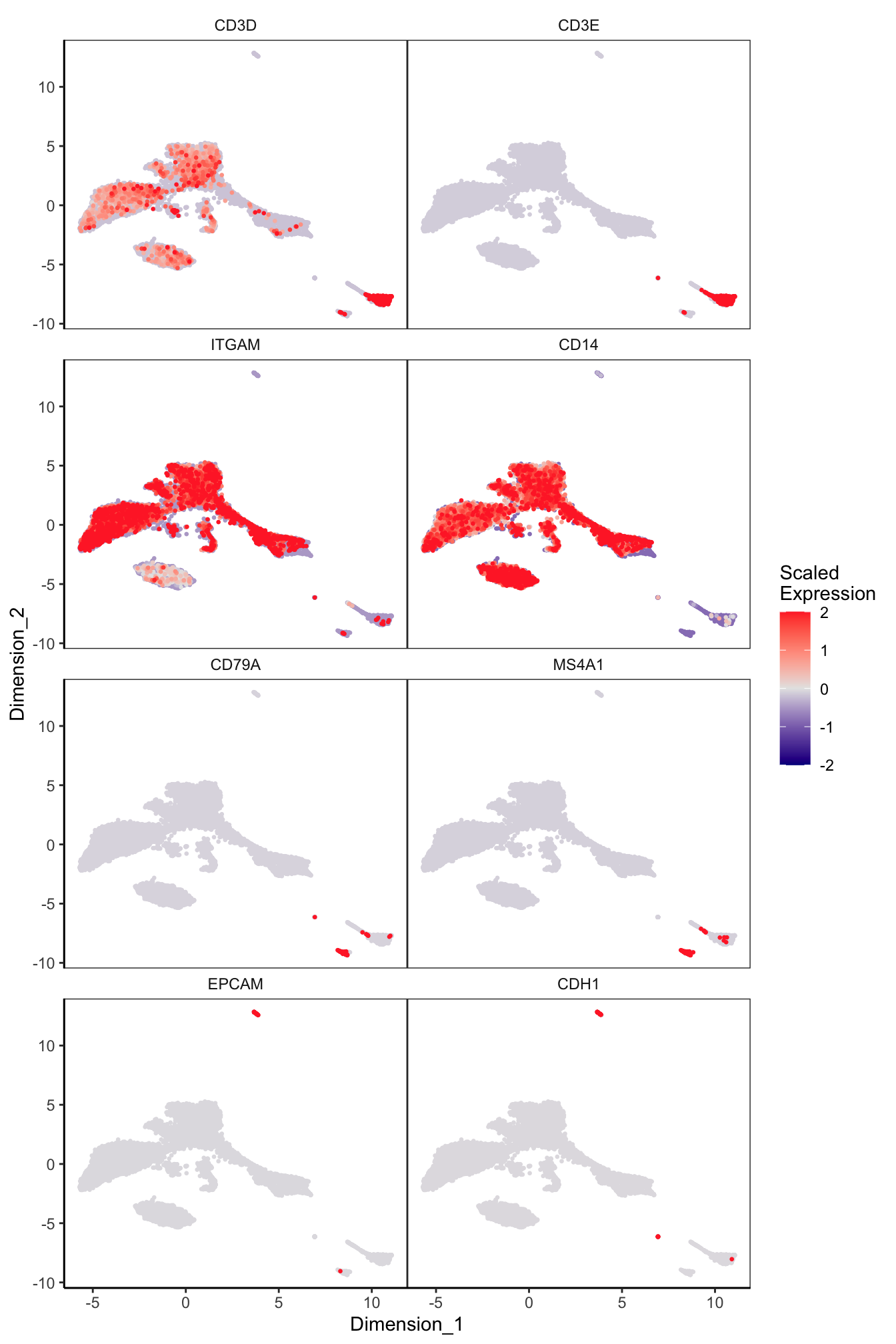
Main cell type markers (after decontX)
p <- lapply(1:length(sceLst), function(i){
sce <- sceLst[[i]]
if(length(levels(sce$Capture)) < 4){
sce_decont <- logNormCounts(sce, assay.type = "decontXcounts")
rownames(sce_decont) <- rowData(sce_decont)$Symbol
umap <- reducedDim(sce_decont, glue::glue("decontX_UMAP"))
plotDimReduceFeature(as.matrix(logcounts(sce_decont)),
dim1 = umap[, 1],
dim2 = umap[, 2],
features = c("CD3D", "CD3E", # T-cells
"ITGAM", "CD14", # Macs
"CD79A", "MS4A1", # B-cells
"EPCAM", "CDH1"), # Epithelial
exactMatch = TRUE,
ncol = 2)
} else {
sce_decont <- logNormCounts(sce, assay.type = "decontXcounts")
rownames(sce_decont) <- rowData(sce_decont)$Symbol
capture_names <- levels(sce_decont$Capture)
p <- lapply(capture_names, function(cn){
umap <- reducedDim(sce_decont, glue::glue("decontX_{cn}_UMAP"))
plotDimReduceFeature(as.matrix(logcounts(sce_decont)),
dim1 = umap[, 1],
dim2 = umap[, 2],
features = c("CD3D", "CD3E", # T-cells
"ITGAM", "CD14", # Macs
"CD79A", "MS4A1", # B-cells
"EPCAM", "CDH1"), # Epithelial
exactMatch = TRUE,
ncol = 2)
})
wrap_plots(p, ncol = 2, guides = "collect") &
theme(legend.position = "bottom",
axis.title = element_text(size = 10),
axis.text = element_text(size = 8))
}
})
p[[1]]
Past versions of unnamed-chunk-7-1.png
Version
Author
Date
8abff7b
Jovana Maksimovic
2024-02-28
[[2]]
Past versions of unnamed-chunk-7-2.png
Version
Author
Date
8abff7b
Jovana Maksimovic
2024-02-28
[[3]]
Past versions of unnamed-chunk-7-3.png
Version
Author
Date
8abff7b
Jovana Maksimovic
2024-02-28
[[4]]
Past versions of unnamed-chunk-7-4.png
Version
Author
Date
8abff7b
Jovana Maksimovic
2024-02-28
[[5]]
Past versions of unnamed-chunk-7-5.png
Version
Author
Date
8abff7b
Jovana Maksimovic
2024-02-28
[[6]]
Past versions of unnamed-chunk-7-6.png
Version
Author
Date
8abff7b
Jovana Maksimovic
2024-02-28
[[7]]
Past versions of unnamed-chunk-7-7.png
Version
Author
Date
8abff7b
Jovana Maksimovic
2024-02-28
Save data
batches <- str_extract(files, "batch[0-6]")
sapply(1:length(sceLst), function(i){
out <- here("data",
paste0("C133_Neeland_", batches[i]),
"data",
"SCEs",
glue::glue("C133_Neeland_{batches[i]}.ambient_removed.SCE.rds"))
if(!file.exists(out)) saveRDS(sceLst[[i]], out)
fs::file_chmod(out, "664")
if(any(str_detect(fs::group_ids()$group_name,
"oshlack_lab"))) fs::file_chown(out,
group_id = "oshlack_lab")
})[[1]]
NULL
[[2]]
NULL
[[3]]
NULL
[[4]]
NULL
[[5]]
NULL
[[6]]
NULL
[[7]]
NULL