Organoids Figures
Last updated: 2018-12-04
workflowr checks: (Click a bullet for more information)-
✔ R Markdown file: up-to-date
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
-
✔ Environment: empty
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
-
✔ Seed:
set.seed(20180730)The command
set.seed(20180730)was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible. -
✔ Session information: recorded
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
-
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.✔ Repository version: 2f21982
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can usewflow_publishorwflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.Ignored files: Ignored: .DS_Store Ignored: .Rhistory Ignored: .Rproj.user/ Ignored: analysis/cache.bak.20181031/ Ignored: analysis/cache.bak/ Ignored: analysis/cache.lind2.20181114/ Ignored: analysis/cache/ Ignored: data/Lindstrom2/ Ignored: data/processed.bak.20181031/ Ignored: data/processed.bak/ Ignored: data/processed.lind2.20181114/ Ignored: packrat/lib-R/ Ignored: packrat/lib-ext/ Ignored: packrat/lib/ Ignored: packrat/src/ Unstaged changes: Modified: output/04D_Organoids_Figures/figure1A.pdf Modified: output/04D_Organoids_Figures/figure1B.pdf Modified: output/04D_Organoids_Figures/figure1C.pdf Modified: output/04D_Organoids_Figures/figure1F.pdf Modified: output/04D_Organoids_Figures/figure1_panel.pdf Modified: output/07D_Combined_Figures/figure2B.pdf Modified: output/07D_Combined_Figures/figure2E.pdf Modified: output/07D_Combined_Figures/figure2F.pdf Modified: output/07D_Combined_Figures/figure2G.pdf Modified: output/07D_Combined_Figures/figure2_panel.pdf Modified: output/07D_Combined_Figures/figure3A.pdf Modified: output/07D_Combined_Figures/figure3B.pdf Modified: output/07D_Combined_Figures/figure3D.pdf Modified: output/07D_Combined_Figures/figure3E.pdf Modified: output/07D_Combined_Figures/figure3F.pdf Modified: output/07D_Combined_Figures/figure3_panel.pdf
Expand here to see past versions:
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 2f21982 | Luke Zappia | 2018-12-04 | Minor updates to figures |
| html | 1b1ce1c | Luke Zappia | 2018-11-23 | Update gene lists for figures |
| html | a1f9f38 | Luke Zappia | 2018-11-23 | Revise figures |
| Rmd | e86d7ce | Luke Zappia | 2018-11-21 | Update organoids figures |
| html | e86d7ce | Luke Zappia | 2018-11-21 | Update organoids figures |
| html | 2354d70 | Luke Zappia | 2018-09-13 | Tidy output files |
| html | a61f9c9 | Luke Zappia | 2018-09-13 | Rebuild site |
| html | ad10b21 | Luke Zappia | 2018-09-13 | Switch to GitHub |
| Rmd | ff4bd7c | Luke Zappia | 2018-09-13 | Rename proximal early nephron clusters |
| Rmd | 22002fe | Luke Zappia | 2018-09-10 | Fix trajectory colours |
| Rmd | 91342d1 | Luke Zappia | 2018-09-10 | Adjust colours and remove shadows |
| Rmd | ebfe6e5 | Luke Zappia | 2018-09-10 | Remove Figure 1E |
# scRNA-seq
library("Seurat")
library("monocle")
# Plotting
library("clustree")
library("cowplot")
# Presentation
library("glue")
library("knitr")
# Parallel
# Paths
library("here")
# Output
# Tidyverse
library("tidyverse")source(here("R/output.R"))orgs.path <- here("data/processed/Organoids_clustered.Rds")
orgs.neph.path <- here("data/processed/Organoids_nephron.Rds")
orgs.neph.cds.path <- here("data/processed/Organoids_trajectory.Rds")
dir.create(here("output", DOCNAME), showWarnings = FALSE)Introduction
In this document we are going to look at all of the organoids analysis results and produce a series of figures for the paper.
if (file.exists(orgs.path)) {
orgs <- read_rds(orgs.path)
} else {
stop("Clustered Organoids dataset is missing. ",
"Please run '04_Organoids_Clustering.Rmd' first.",
call. = FALSE)
}if (file.exists(orgs.neph.path)) {
orgs.neph <- read_rds(orgs.neph.path)
} else {
stop("Clustered Organoids nephron dataset is missing. ",
"Please run '04B_Organoids_Nephron.Rmd' first.",
call. = FALSE)
}if (file.exists(orgs.neph.cds.path)) {
orgs.neph.cds <- read_rds(orgs.neph.cds.path)
} else {
stop("Organoids nephron trajectory dataset is missing. ",
"Please run '04C_Organoids_Trajectory.Rmd' first.",
call. = FALSE)
}Figure 1A
plot.data <- orgs %>%
GetDimReduction("tsne", slot = "cell.embeddings") %>%
data.frame() %>%
rownames_to_column("Cell") %>%
mutate(Cluster = orgs@ident) %>%
group_by(Cluster)
lab.data <- plot.data %>%
group_by(Cluster) %>%
summarise(tSNE_1 = mean(tSNE_1),
tSNE_2 = mean(tSNE_2)) %>%
mutate(Label = paste0("O", Cluster))
clust.labs <- c(
"O0 (Stroma)\nTAGLN, ACTA2, MGP\ncardiovascular system development",
"O1 (Stroma)\nMAB21L2, CXCL14, PRRX1\ncartilage development",
"O2 (Podocyte)\nPODXL, NPHS2, TCF21\nrenal filtration cell differentiation",
"O3 (Stroma)\nDLK1, GATA3, IGFBP5\nwound healing",
"O4 (Cell cycle)\nHIST1H4C, PCLAF, TYMS\nDNA conformation change",
"O5 (Endothelium)\nCLDN5, PECAM1, KDR\ncardiovascular system development",
"O6 (Cell cycle)\nCENPF, HMGB2, UBE2C\nmitotic cell cycle processes",
"O7 (Stroma)\nCOL2A1, COL9A3, CNMD\nextracellular matrix organisation",
"O8 (Glial)\nFABP7, TTYH1, SOX2\nbrain development",
"O9 (Epithelium)\nPAX2, PAX8, KRT19\nreg. of nephron tubule differentiation",
"O10 (Muscle progenitor)\nMYOG, MYOD1\nmuscle filament sliding",
"O11 (Neural progenitor)\nHES6, STMN2\ngeneration of neurons",
"O12 (Endothelium)\nGNG11, CALM1\nnegative reg. of cation channel activity"
)
f1A <- ggplot(plot.data, aes(x = tSNE_1, y = tSNE_2, colour = Cluster)) +
geom_point() +
geom_text(data = lab.data, aes(label = Label), colour = "black", size = 6) +
scale_colour_discrete(labels = clust.labs) +
guides(colour = guide_legend(ncol = 2, override.aes = list(size = 16),
label.theme = element_text(size = 12),
keyheight = 4)) +
theme_cowplot() +
theme(legend.title = element_blank())
ggsave(here("output", DOCNAME, "figure1A.png"), f1A,
height = 8, width = 10)
ggsave(here("output", DOCNAME, "figure1A.pdf"), f1A,
height = 8, width = 10)
f1A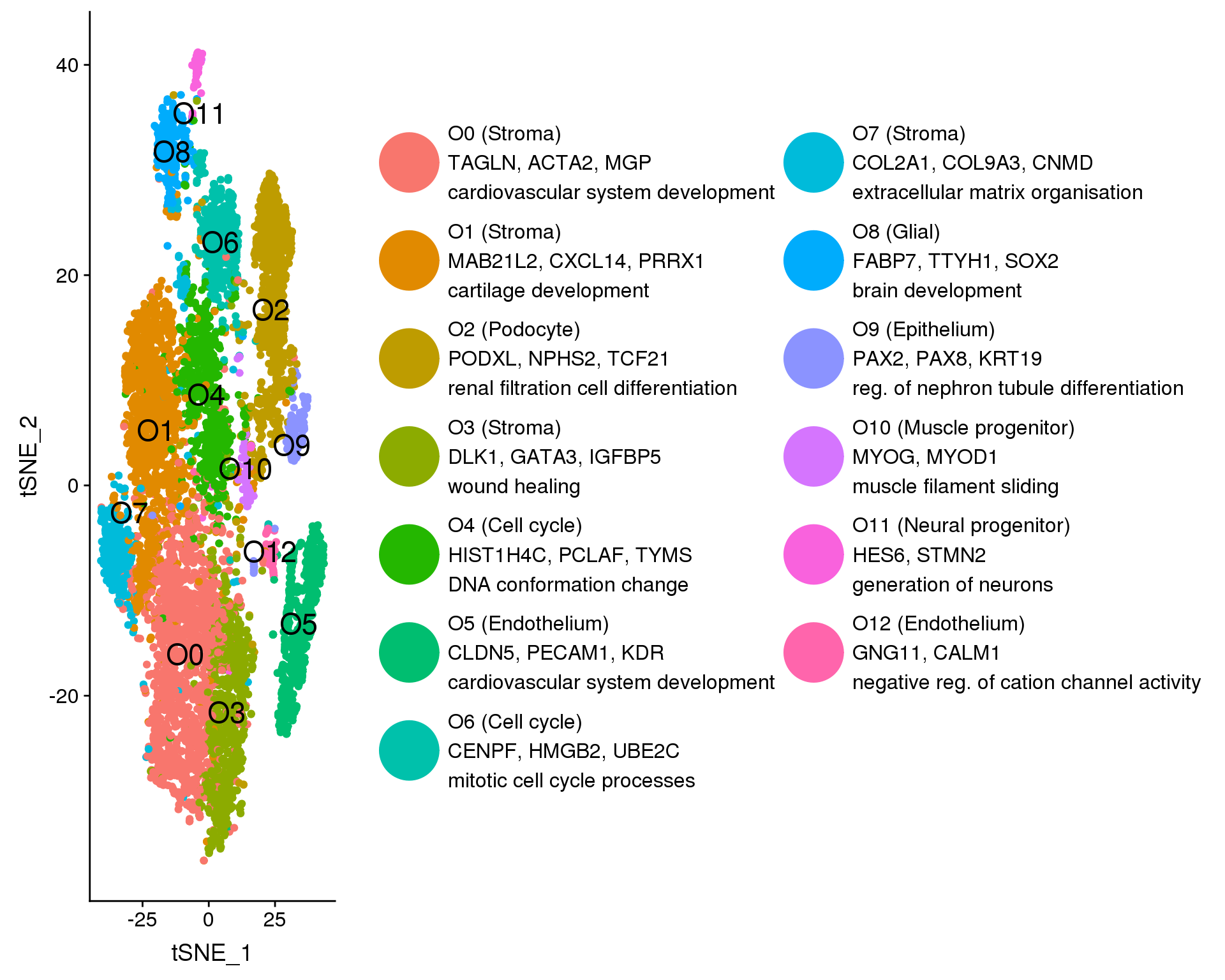
Figure 1B
plot.data <- orgs.neph %>%
GetDimReduction("tsne", slot = "cell.embeddings") %>%
data.frame() %>%
rownames_to_column("Cell") %>%
mutate(Cluster = orgs.neph@ident) %>%
group_by(Cluster)
lab.data <- plot.data %>%
group_by(Cluster) %>%
summarise(tSNE_1 = mean(tSNE_1),
tSNE_2 = mean(tSNE_2)) %>%
mutate(Label = paste0("ON", Cluster))
clust.labs <- c(
"ON0 (Podocyte)\nTCF21, PODXL, VEGFA, NPHS1, PTPRO",
"ON1 (Podocyte precursor)\nCTGF, OLFM3, MAFB, NPHS1",
"ON2 (Nephron progenitor)\nDAPL1, LYPD1, SIX1, CRABP2",
"ON3 (Proximal precursor)\nIGFBP7, FXYD2, CDH6, HNF1B",
"ON4 (Distal precursor)\nEPCAM, EMX2, SPP1, MAL, PAX2"
)
f1B <- ggplot(plot.data, aes(x = tSNE_1, y = tSNE_2, colour = Cluster)) +
geom_point(size = 3) +
geom_text(data = lab.data, aes(label = Label), colour = "black", size = 6) +
#scale_color_brewer(palette = "Set1", labels = clust.labs) +
scale_color_discrete(labels = clust.labs) +
guides(colour = guide_legend(ncol = 2, override.aes = list(size = 12),
label.theme = element_text(size = 12),
keyheight = 3)) +
theme_cowplot() +
theme(legend.position = "bottom",
legend.title = element_blank(),
legend.justification = "center")
ggsave(here("output", DOCNAME, "figure1B.png"), f1B,
height = 8, width = 10)
ggsave(here("output", DOCNAME, "figure1B.pdf"), f1B,
height = 8, width = 10)
f1B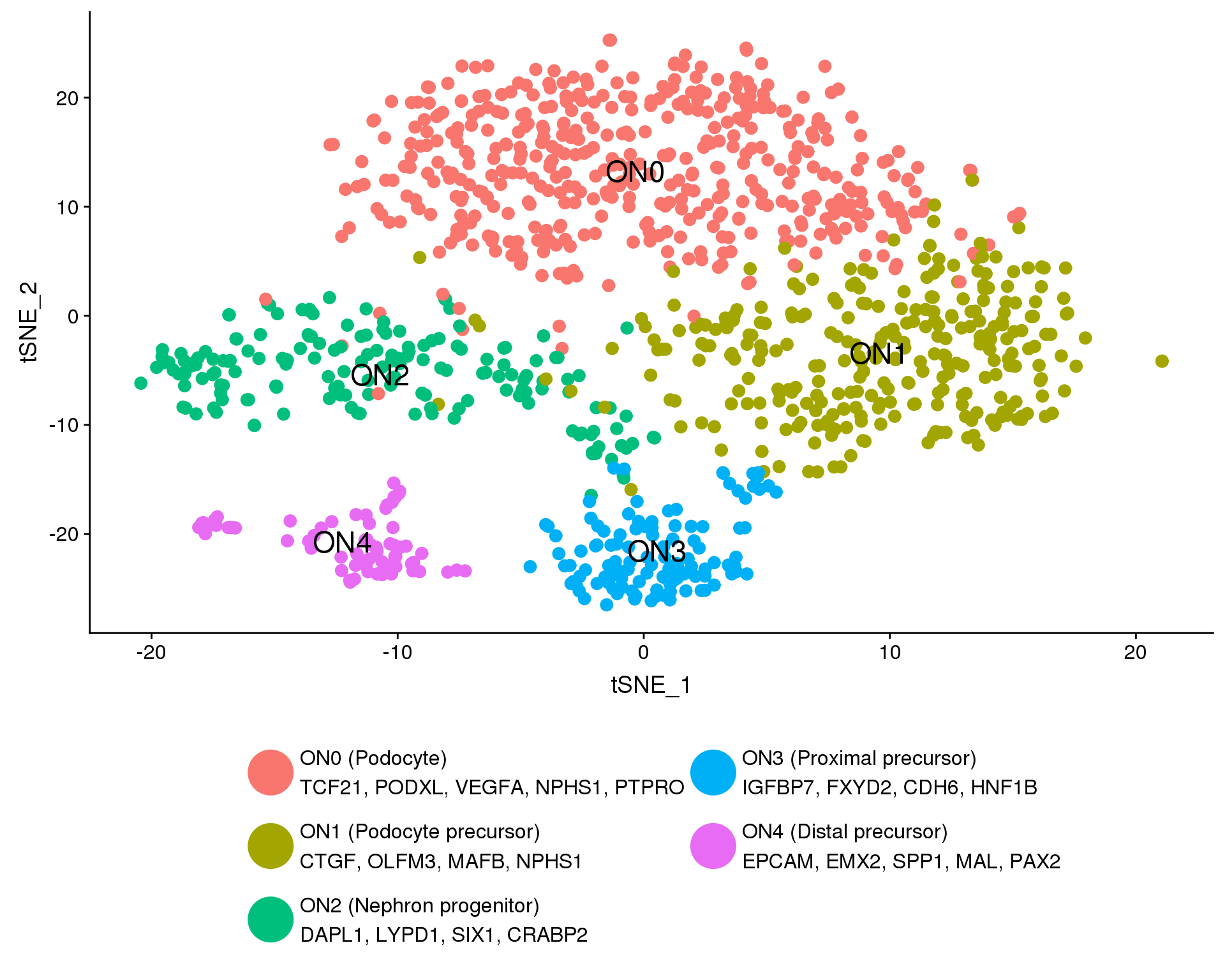
Figure 1C
clust.labs <- c(
"ON0 (Podocyte)",
"ON1 (Proximal early nephron)",
"ON2 (Nephron progenitor)",
"ON3 (Proximal precursor)",
"ON4 (Distal precursor)"
)
f1C <- plot_cell_trajectory(orgs.neph.cds,
color_by = "NephCluster", cell_size = 2) +
scale_color_discrete(labels = clust.labs) +
guides(colour = guide_legend(nrow = 2, override.aes = list(size = 8),
label.theme = element_text(size = 11))) +
theme_cowplot() +
theme(legend.position = "bottom",
legend.title = element_blank())
ggsave(here("output", DOCNAME, "figure1C.png"), f1C,
height = 8, width = 10)
ggsave(here("output", DOCNAME, "figure1C.pdf"), f1C,
height = 8, width = 10)
f1C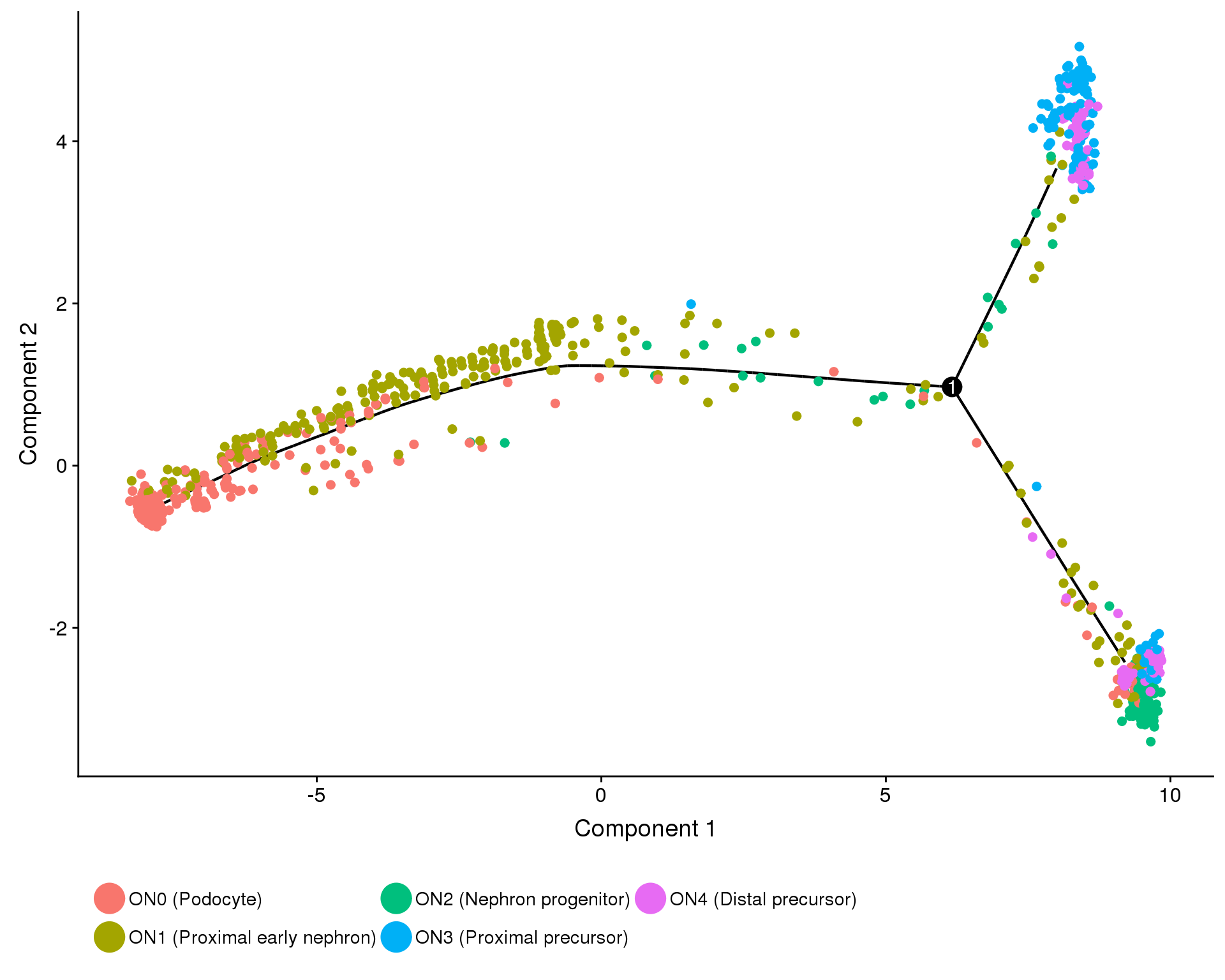
Figure 1D
annot.data <- tribble(
~Text, ~x, ~y, ~State,
"State 1", 6.0, -2.5, "1",
"State 2", -6.5, 2.2, "2",
"State 3", 5.0, 4.5, "3"
)
f1D <- plot_cell_trajectory(orgs.neph.cds,
color_by = "State", cell_size = 2) +
geom_text(data = annot.data,
aes(x = x, y = y, label = Text, colour = State),
size = 6) +
guides(colour = guide_legend(nrow = 2, override.aes = list(size = 8),
label.theme = element_text(size = 11))) +
theme_cowplot() +
theme(legend.position = "none")
ggsave(here("output", DOCNAME, "figure1D.png"), f1D,
height = 8, width = 10)
ggsave(here("output", DOCNAME, "figure1D.pdf"), f1D,
height = 8, width = 10)
f1D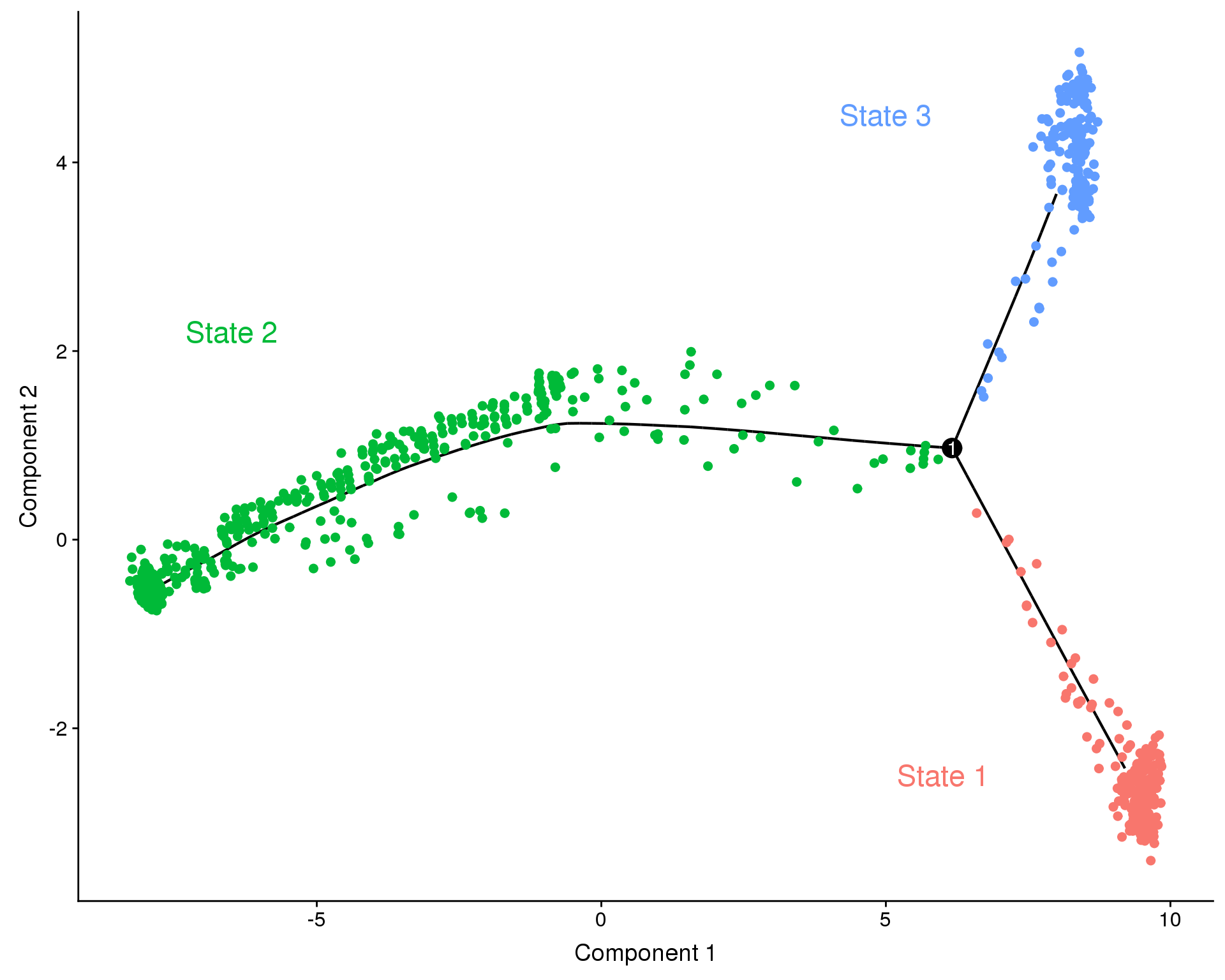
Expand here to see past versions of fig-1D-1.png:
| Version | Author | Date |
|---|---|---|
| ad10b21 | Luke Zappia | 2018-09-13 |
Figure 1E
genes <- c("PODXL", "NPHS1", "DAPL1", "SIX1", "MAL", "HNF1B")
f1E <- plot_genes_branched_pseudotime(
orgs.neph.cds[genes, ],
branch_point = 1,
branch_labels = c("Nephron", "Tubule"),
ncol = 2,
panel_order = genes,
color_by = "State",
trend_formula = "~ sm.ns(Pseudotime, df=2) * Branch"
) +
theme(legend.position = "bottom",
legend.justification = "center")
ggsave(here("output", DOCNAME, "figure1E.png"), f1E,
height = 8, width = 10)
ggsave(here("output", DOCNAME, "figure1E.pdf"), f1E,
height = 8, width = 10)
f1E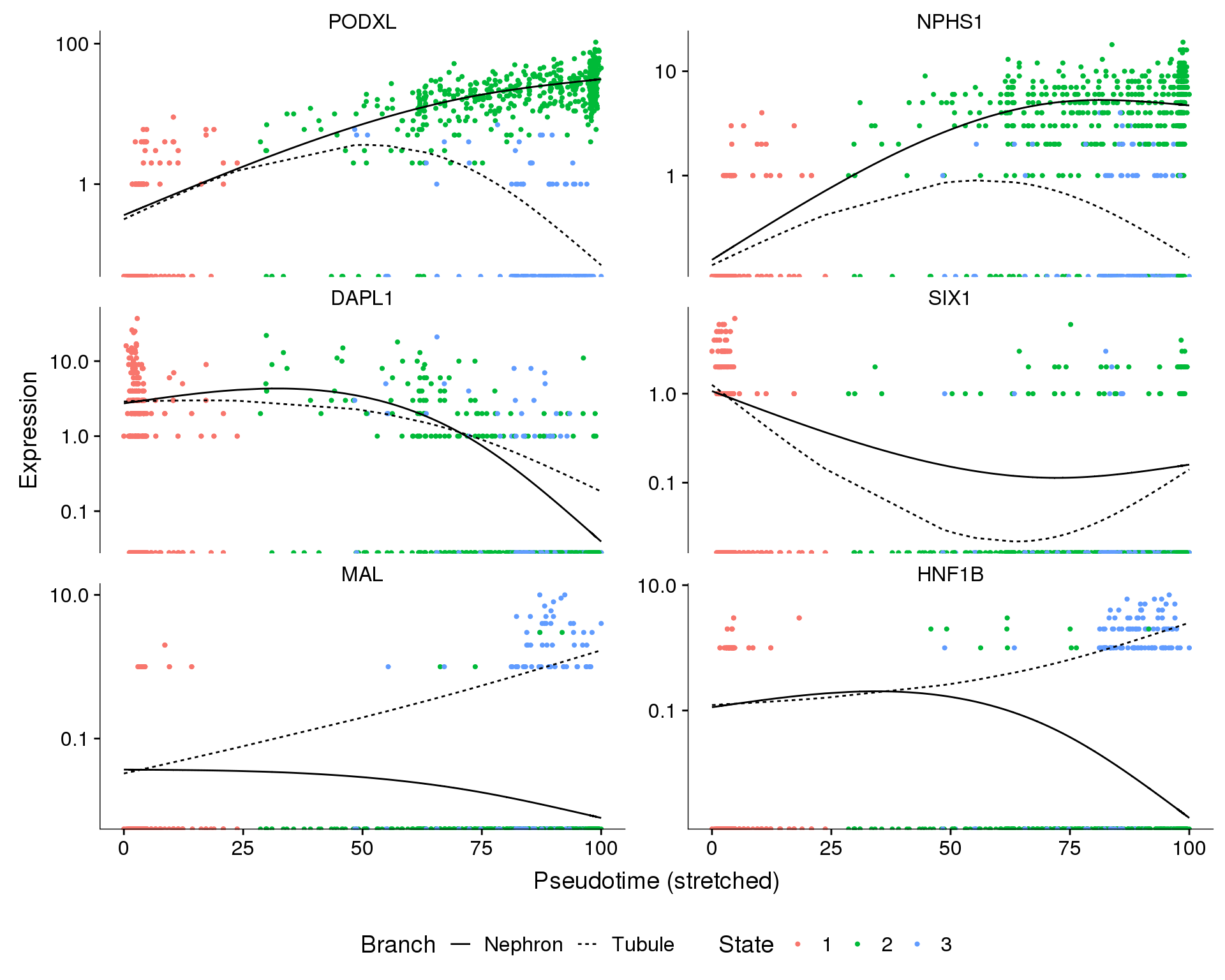
Figure 1F
genes <- c("TCF21", "PODXL", "VEGFA", "NPHS1", "PTPRO", "CTGF", "OLFM3",
"MAFB", "DAPL1", "LYPD1", "SIX1", "CRABP2", "IGFBP7", "FXYD2",
"CDH6", "HNF1B", "EPCAM", "EMX2", "SPP1", "MAL", "PAX2")
gene.groups <- factor(c(rep("Podocyte", 5),
rep("Podocyte precursor", 3),
rep("Nephron progenitor", 4),
rep("Proximal precursor", 4),
rep("Distal precursor", 5)),
levels = c("Podocyte", "Podocyte precursor",
"Nephron progenitor", "Proximal precursor",
"Distal precursor"))
names(gene.groups) <- genes
clust.labs <- c(
"ON0",
"ON1",
"ON2",
"ON3",
"ON4"
)
plot.data <- data.frame(FetchData(orgs.neph, vars.all = genes)) %>%
rownames_to_column("Cell") %>%
mutate(Cluster = orgs.neph@ident) %>%
gather(key = "Gene", value = "Expr", -Cell, -Cluster) %>%
group_by(Cluster, Gene) %>%
summarize(AvgExpr = mean(expm1(Expr)),
PctExpr = Seurat:::PercentAbove(Expr, threshold = 0) * 100) %>%
group_by(Gene) %>%
mutate(AvgExprScale = scale(AvgExpr)) %>%
mutate(AvgExprScale = Seurat::MinMax(AvgExprScale,
max = 2.5, min = -2.5)) %>%
ungroup() %>%
mutate(Group = gene.groups[Gene]) %>%
mutate(Gene = factor(Gene, levels = genes))
f1F <- ggplot(plot.data,
aes(x = Gene, y = Cluster, size = PctExpr,
alpha = AvgExprScale)) +
geom_point(colour = "#00ADEF") +
scale_radius(range = c(0, 10)) +
scale_y_discrete(labels = clust.labs) +
facet_grid(~ Group, scales = "free_x") +
theme(axis.title.x = element_blank(),
axis.title.y = element_blank(),
axis.text.x = element_text(angle = 90, vjust = 0.5, hjust = 1),
panel.spacing = unit(x = 1, units = "lines"),
strip.background = element_blank(),
strip.placement = "outside",
legend.position = "none")
ggsave(here("output", DOCNAME, "figure1F.png"), f1F,
height = 8, width = 10)
ggsave(here("output", DOCNAME, "figure1F.pdf"), f1F,
height = 8, width = 10)
f1F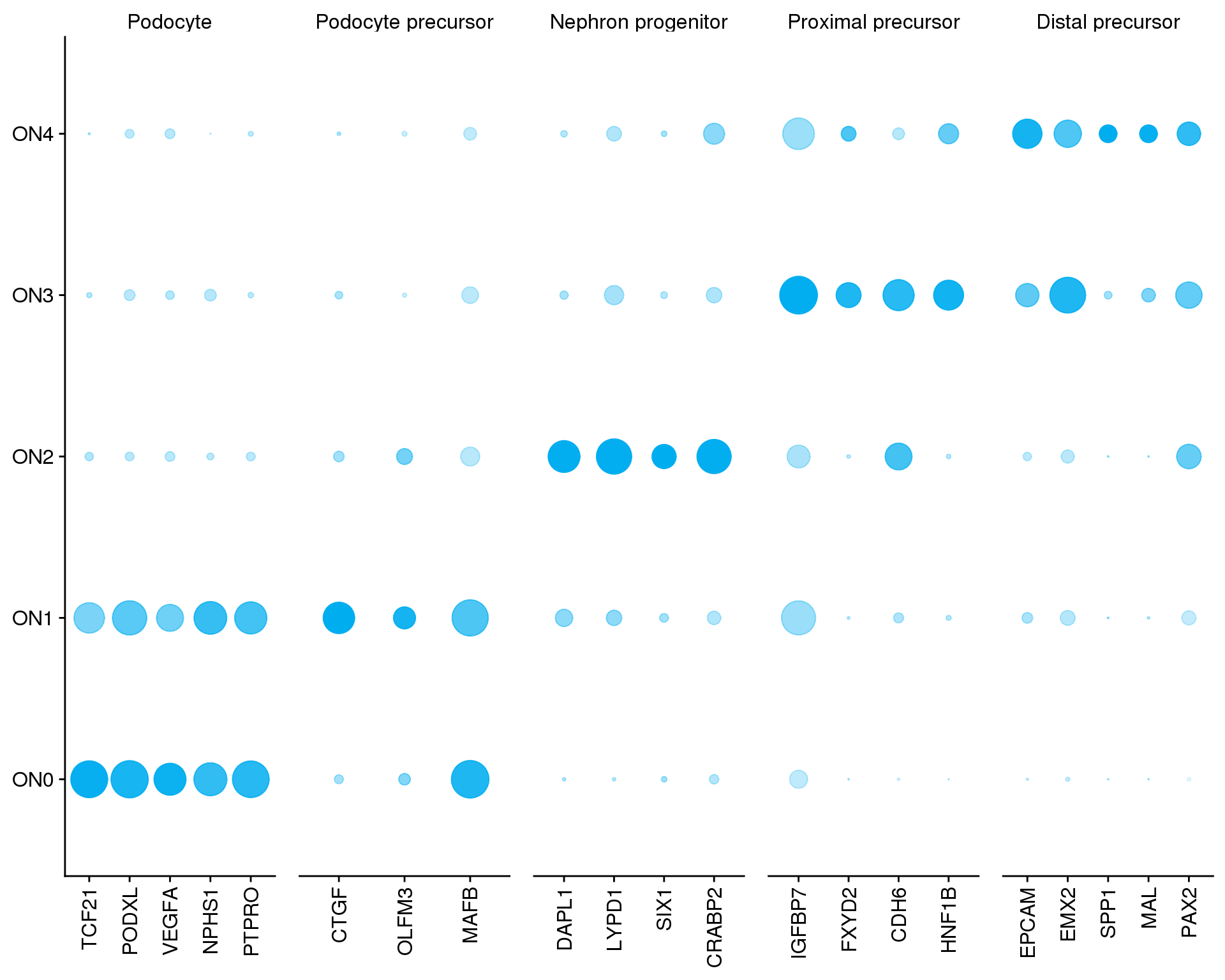
Figure 1 Panel
p1 <- plot_grid(f1C + theme(legend.position = "none"), f1D,
nrow = 1, labels = c("C", "D"),
label_size = 20)
p2 <- plot_grid(p1, f1E,
nrow = 2, labels = c("", "E"),
label_size = 20)
p3 <- plot_grid(f1B, p2,
nrow = 1, labels = c("B", ""),
label_size = 20)
panel <- plot_grid(f1A, p3, f1F,
nrow = 3, labels = c("A", "", "F"),
rel_heights = c(0.8, 1, 0.5),
label_size = 20)
ggsave(here("output", DOCNAME, "figure1_panel.png"), panel,
height = 18, width = 16)
ggsave(here("output", DOCNAME, "figure1_panel.pdf"), panel,
height = 18, width = 16)
panel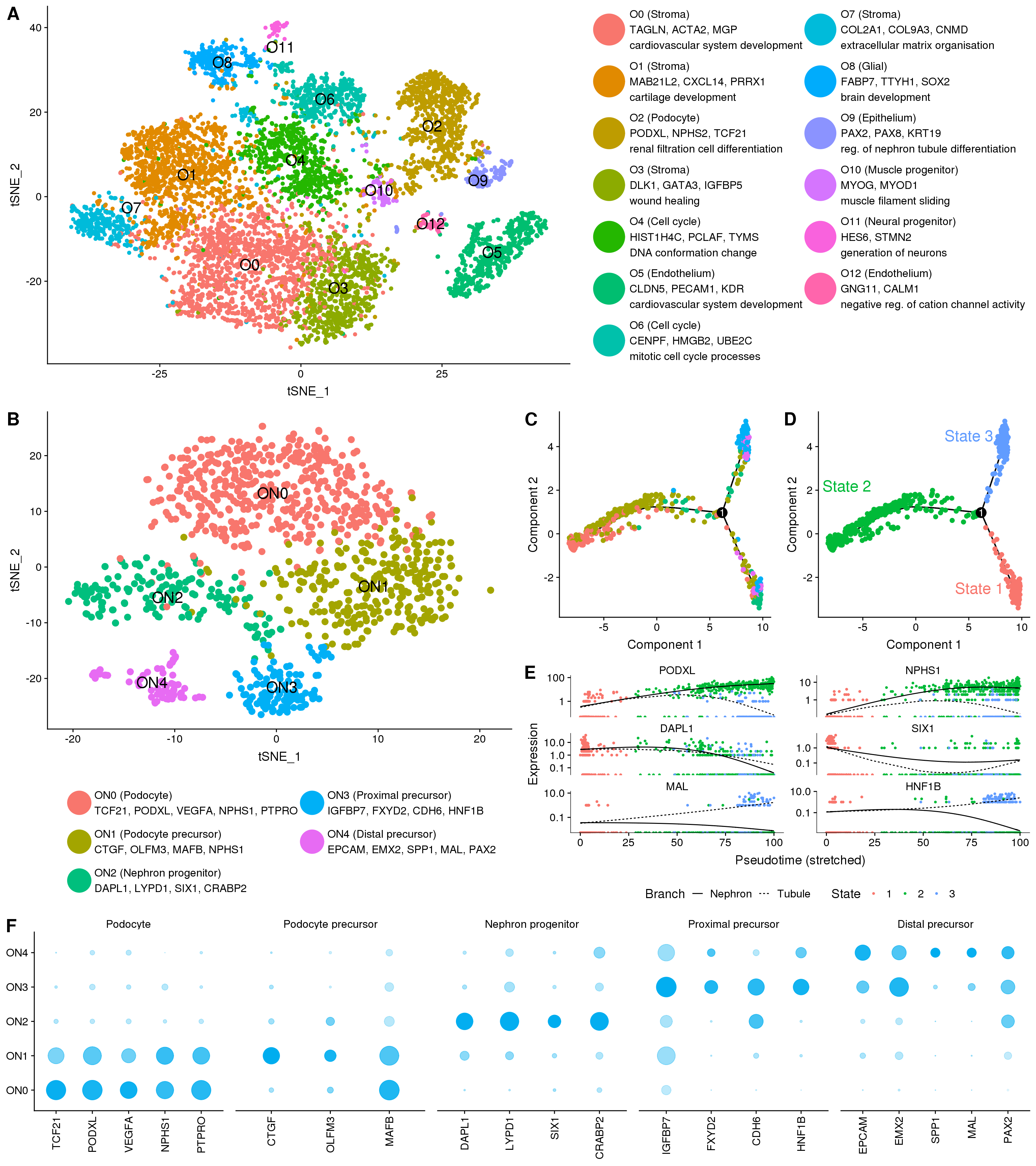
Expand here to see past versions of fig1-panel-1.png:
| Version | Author | Date |
|---|---|---|
| 1b1ce1c | Luke Zappia | 2018-11-23 |
| a1f9f38 | Luke Zappia | 2018-11-23 |
| e86d7ce | Luke Zappia | 2018-11-21 |
| ad10b21 | Luke Zappia | 2018-09-13 |
f1A <- clustree(orgs, node_size_range = c(6, 16), node_text_size = 5,
edge_width = 2)
leg <- {f1A + guides(colour = FALSE,
edge_alpha = guide_legend(title = "In proportion",
title.position = "top",
label.position = "top",
override.aes = list(edge_width = 2),
keywidth = 3),
edge_colour = guide_edge_colourbar(title = "Cell count",
title.position = "top",
barwidth = 15,
barheight = 2.5),
size = guide_legend(title = "Cluster size",
title.position = "top",
label.position = "top")) +
theme(legend.position = "bottom")} %>%
get_legend()
f1A <- f1A +
annotate("rect", xmin = -9, xmax = 6, ymin = 3.6, ymax = 4.4,
alpha = 0, colour = "#00ADEF", size = 1.5) +
scale_colour_viridis_d(option = "inferno", begin = 0.4, end = 0.9) +
guides(size = FALSE, edge_alpha = FALSE, edge_colour = FALSE,
colour = guide_legend(override.aes = list(size = 8),
keyheight = 3,
title = "Clustering resolution",
label.theme = element_text(size = 12),
title.position = "left",
title.theme = element_text(size = 16,
angle = 90,
hjust = 0.5))) +
theme(legend.position = "left")
f1A <- plot_grid(f1A, leg, ncol = 1, rel_heights = c(1, 0.2))
ggsave(here("output", DOCNAME, "figure1A.png"), f1A,
height = 8, width = 10)
ggsave(here("output", DOCNAME, "figure1A.pdf"), f1A,
height = 8, width = 10)
f1ASummary
Output files
This table describes the output files produced by this document. Right click and Save Link As… to download the results.
kable(data.frame(
File = c(
glue("[figure1A.png]({getDownloadURL('figure1A.png', DOCNAME)})"),
glue("[figure1A.pdf]({getDownloadURL('figure1A.pdf', DOCNAME)})"),
glue("[figure1B.png]({getDownloadURL('figure1B.png', DOCNAME)})"),
glue("[figure1B.pdf]({getDownloadURL('figure1B.pdf', DOCNAME)})"),
glue("[figure1C.png]({getDownloadURL('figure1C.png', DOCNAME)})"),
glue("[figure1C.pdf]({getDownloadURL('figure1C.pdf', DOCNAME)})"),
glue("[figure1D.png]({getDownloadURL('figure1D.png', DOCNAME)})"),
glue("[figure1D.pdf]({getDownloadURL('figure1D.pdf', DOCNAME)})"),
glue("[figure1E.png]({getDownloadURL('figure1D.png', DOCNAME)})"),
glue("[figure1E.pdf]({getDownloadURL('figure1D.pdf', DOCNAME)})"),
glue("[figure1F.png]({getDownloadURL('figure1D.png', DOCNAME)})"),
glue("[figure1F.pdf]({getDownloadURL('figure1D.pdf', DOCNAME)})"),
glue("[figure1_panel.png]",
"({getDownloadURL('figure1_panel.png', DOCNAME)})"),
glue("[figure1_panel.pdf]",
"({getDownloadURL('figure1_panel.pdf', DOCNAME)})")
),
Description = c(
"Figure 1A in PNG format",
"Figure 1A in PDF format",
"Figure 1B in PNG format",
"Figure 1B in PDF format",
"Figure 1C in PNG format",
"Figure 1C in PDF format",
"Figure 1D in PNG format",
"Figure 1D in PDF format",
"Figure 1E in PNG format",
"Figure 1E in PDF format",
"Figure 1F in PNG format",
"Figure 1F in PDF format",
"Figure panel in PNG format",
"Figure panel in PDF format"
)
))| File | Description |
|---|---|
| figure1A.png | Figure 1A in PNG format |
| figure1A.pdf | Figure 1A in PDF format |
| figure1B.png | Figure 1B in PNG format |
| figure1B.pdf | Figure 1B in PDF format |
| figure1C.png | Figure 1C in PNG format |
| figure1C.pdf | Figure 1C in PDF format |
| figure1D.png | Figure 1D in PNG format |
| figure1D.pdf | Figure 1D in PDF format |
| figure1E.png | Figure 1E in PNG format |
| figure1E.pdf | Figure 1E in PDF format |
| figure1F.png | Figure 1F in PNG format |
| figure1F.pdf | Figure 1F in PDF format |
| figure1_panel.png | Figure panel in PNG format |
| figure1_panel.pdf | Figure panel in PDF format |
Session information
devtools::session_info() setting value
version R version 3.5.0 (2018-04-23)
system x86_64, linux-gnu
ui X11
language (EN)
collate en_US.UTF-8
tz Australia/Melbourne
date 2018-12-04
package * version date source
abind 1.4-5 2016-07-21 cran (@1.4-5)
acepack 1.4.1 2016-10-29 cran (@1.4.1)
ape 5.1 2018-04-04 cran (@5.1)
assertthat 0.2.0 2017-04-11 CRAN (R 3.5.0)
backports 1.1.2 2017-12-13 CRAN (R 3.5.0)
base * 3.5.0 2018-06-18 local
base64enc 0.1-3 2015-07-28 CRAN (R 3.5.0)
bibtex 0.4.2 2017-06-30 cran (@0.4.2)
bindr 0.1.1 2018-03-13 cran (@0.1.1)
bindrcpp 0.2.2 2018-03-29 cran (@0.2.2)
Biobase * 2.40.0 2018-07-30 Bioconductor
BiocGenerics * 0.26.0 2018-07-30 Bioconductor
bitops 1.0-6 2013-08-17 cran (@1.0-6)
broom 0.5.0 2018-07-17 cran (@0.5.0)
caret 6.0-80 2018-05-26 cran (@6.0-80)
caTools 1.17.1.1 2018-07-20 cran (@1.17.1.)
cellranger 1.1.0 2016-07-27 CRAN (R 3.5.0)
checkmate 1.8.5 2017-10-24 cran (@1.8.5)
class 7.3-14 2015-08-30 CRAN (R 3.5.0)
cli 1.0.0 2017-11-05 CRAN (R 3.5.0)
cluster 2.0.7-1 2018-04-13 CRAN (R 3.5.0)
clustree * 0.2.2.9000 2018-08-01 Github (lazappi/clustree@66a865b)
codetools 0.2-15 2016-10-05 CRAN (R 3.5.0)
colorspace 1.3-2 2016-12-14 cran (@1.3-2)
combinat 0.0-8 2012-10-29 CRAN (R 3.5.0)
compiler 3.5.0 2018-06-18 local
cowplot * 0.9.3 2018-07-15 cran (@0.9.3)
crayon 1.3.4 2017-09-16 CRAN (R 3.5.0)
CVST 0.2-2 2018-05-26 cran (@0.2-2)
data.table 1.11.4 2018-05-27 cran (@1.11.4)
datasets * 3.5.0 2018-06-18 local
ddalpha 1.3.4 2018-06-23 cran (@1.3.4)
DDRTree * 0.1.5 2017-04-30 CRAN (R 3.5.0)
densityClust 0.3 2017-10-24 CRAN (R 3.5.0)
DEoptimR 1.0-8 2016-11-19 cran (@1.0-8)
devtools 1.13.6 2018-06-27 CRAN (R 3.5.0)
diffusionMap 1.1-0.1 2018-07-21 cran (@1.1-0.1)
digest 0.6.15 2018-01-28 CRAN (R 3.5.0)
dimRed 0.1.0 2017-05-04 cran (@0.1.0)
diptest 0.75-7 2016-12-05 cran (@0.75-7)
docopt 0.6 2018-08-03 CRAN (R 3.5.0)
doSNOW 1.0.16 2017-12-13 cran (@1.0.16)
dplyr * 0.7.6 2018-06-29 cran (@0.7.6)
DRR 0.0.3 2018-01-06 cran (@0.0.3)
dtw 1.20-1 2018-05-18 cran (@1.20-1)
evaluate 0.10.1 2017-06-24 CRAN (R 3.5.0)
fastICA 1.2-1 2017-06-12 CRAN (R 3.5.0)
fitdistrplus 1.0-9 2017-03-24 cran (@1.0-9)
flexmix 2.3-14 2017-04-28 cran (@2.3-14)
FNN 1.1 2013-07-31 cran (@1.1)
forcats * 0.3.0 2018-02-19 CRAN (R 3.5.0)
foreach 1.4.4 2017-12-12 cran (@1.4.4)
foreign 0.8-71 2018-07-20 CRAN (R 3.5.0)
Formula 1.2-3 2018-05-03 cran (@1.2-3)
fpc 2.1-11.1 2018-07-20 cran (@2.1-11.)
gbRd 0.4-11 2012-10-01 cran (@0.4-11)
gdata 2.18.0 2017-06-06 cran (@2.18.0)
geometry 0.3-6 2015-09-09 cran (@0.3-6)
ggforce 0.1.3 2018-07-07 CRAN (R 3.5.0)
ggplot2 * 3.0.0 2018-07-03 cran (@3.0.0)
ggraph * 1.0.2 2018-07-07 CRAN (R 3.5.0)
ggrepel 0.8.0 2018-05-09 CRAN (R 3.5.0)
ggridges 0.5.0 2018-04-05 cran (@0.5.0)
git2r 0.21.0 2018-01-04 CRAN (R 3.5.0)
glue * 1.3.0 2018-07-17 cran (@1.3.0)
gower 0.1.2 2017-02-23 cran (@0.1.2)
gplots 3.0.1 2016-03-30 cran (@3.0.1)
graphics * 3.5.0 2018-06-18 local
grDevices * 3.5.0 2018-06-18 local
grid 3.5.0 2018-06-18 local
gridExtra 2.3 2017-09-09 cran (@2.3)
gtable 0.2.0 2016-02-26 cran (@0.2.0)
gtools 3.8.1 2018-06-26 cran (@3.8.1)
haven 1.1.2 2018-06-27 CRAN (R 3.5.0)
here * 0.1 2017-05-28 CRAN (R 3.5.0)
Hmisc 4.1-1 2018-01-03 cran (@4.1-1)
hms 0.4.2 2018-03-10 CRAN (R 3.5.0)
HSMMSingleCell 0.114.0 2018-08-28 Bioconductor
htmlTable 1.12 2018-05-26 cran (@1.12)
htmltools 0.3.6 2017-04-28 CRAN (R 3.5.0)
htmlwidgets 1.2 2018-04-19 cran (@1.2)
httr 1.3.1 2017-08-20 CRAN (R 3.5.0)
ica 1.0-2 2018-05-24 cran (@1.0-2)
igraph 1.2.2 2018-07-27 cran (@1.2.2)
ipred 0.9-6 2017-03-01 cran (@0.9-6)
irlba * 2.3.2 2018-01-11 cran (@2.3.2)
iterators 1.0.10 2018-07-13 cran (@1.0.10)
jsonlite 1.5 2017-06-01 CRAN (R 3.5.0)
kernlab 0.9-26 2018-04-30 cran (@0.9-26)
KernSmooth 2.23-15 2015-06-29 CRAN (R 3.5.0)
knitr * 1.20 2018-02-20 CRAN (R 3.5.0)
lars 1.2 2013-04-24 cran (@1.2)
lattice 0.20-35 2017-03-25 CRAN (R 3.5.0)
latticeExtra 0.6-28 2016-02-09 cran (@0.6-28)
lava 1.6.2 2018-07-02 cran (@1.6.2)
lazyeval 0.2.1 2017-10-29 cran (@0.2.1)
limma 3.36.2 2018-06-21 Bioconductor
lmtest 0.9-36 2018-04-04 cran (@0.9-36)
lubridate 1.7.4 2018-04-11 cran (@1.7.4)
magic 1.5-8 2018-01-26 cran (@1.5-8)
magrittr 1.5 2014-11-22 CRAN (R 3.5.0)
MASS 7.3-50 2018-04-30 CRAN (R 3.5.0)
Matrix * 1.2-14 2018-04-09 CRAN (R 3.5.0)
matrixStats 0.54.0 2018-07-23 CRAN (R 3.5.0)
mclust 5.4.1 2018-06-27 cran (@5.4.1)
memoise 1.1.0 2017-04-21 CRAN (R 3.5.0)
metap 1.0 2018-07-25 cran (@1.0)
methods * 3.5.0 2018-06-18 local
mixtools 1.1.0 2017-03-10 cran (@1.1.0)
ModelMetrics 1.1.0 2016-08-26 cran (@1.1.0)
modelr 0.1.2 2018-05-11 CRAN (R 3.5.0)
modeltools 0.2-22 2018-07-16 cran (@0.2-22)
monocle * 2.8.0 2018-08-28 Bioconductor
munsell 0.5.0 2018-06-12 cran (@0.5.0)
mvtnorm 1.0-8 2018-05-31 cran (@1.0-8)
nlme 3.1-137 2018-04-07 CRAN (R 3.5.0)
nnet 7.3-12 2016-02-02 CRAN (R 3.5.0)
parallel * 3.5.0 2018-06-18 local
pbapply 1.3-4 2018-01-10 cran (@1.3-4)
pheatmap 1.0.10 2018-05-19 CRAN (R 3.5.0)
pillar 1.3.0 2018-07-14 cran (@1.3.0)
pkgconfig 2.0.1 2017-03-21 cran (@2.0.1)
pls 2.6-0 2016-12-18 cran (@2.6-0)
plyr 1.8.4 2016-06-08 cran (@1.8.4)
png 0.1-7 2013-12-03 cran (@0.1-7)
prabclus 2.2-6 2015-01-14 cran (@2.2-6)
prodlim 2018.04.18 2018-04-18 cran (@2018.04)
proxy 0.4-22 2018-04-08 cran (@0.4-22)
purrr * 0.2.5 2018-05-29 cran (@0.2.5)
qlcMatrix 0.9.7 2018-04-20 CRAN (R 3.5.0)
R.methodsS3 1.7.1 2016-02-16 CRAN (R 3.5.0)
R.oo 1.22.0 2018-04-22 CRAN (R 3.5.0)
R.utils 2.6.0 2017-11-05 CRAN (R 3.5.0)
R6 2.2.2 2017-06-17 CRAN (R 3.5.0)
ranger 0.10.1 2018-06-04 cran (@0.10.1)
RANN 2.6 2018-07-16 cran (@2.6)
RColorBrewer 1.1-2 2014-12-07 cran (@1.1-2)
Rcpp 0.12.18 2018-07-23 cran (@0.12.18)
RcppRoll 0.3.0 2018-06-05 cran (@0.3.0)
Rdpack 0.8-0 2018-05-24 cran (@0.8-0)
readr * 1.1.1 2017-05-16 CRAN (R 3.5.0)
readxl 1.1.0 2018-04-20 CRAN (R 3.5.0)
recipes 0.1.3 2018-06-16 cran (@0.1.3)
reshape2 1.4.3 2017-12-11 cran (@1.4.3)
reticulate 1.9 2018-07-06 cran (@1.9)
rlang 0.2.1 2018-05-30 CRAN (R 3.5.0)
rmarkdown 1.10.2 2018-07-30 Github (rstudio/rmarkdown@18207b9)
robustbase 0.93-2 2018-07-27 cran (@0.93-2)
ROCR 1.0-7 2015-03-26 cran (@1.0-7)
rpart 4.1-13 2018-02-23 CRAN (R 3.5.0)
rprojroot 1.3-2 2018-01-03 CRAN (R 3.5.0)
rstudioapi 0.7 2017-09-07 CRAN (R 3.5.0)
Rtsne 0.13 2017-04-14 cran (@0.13)
rvest 0.3.2 2016-06-17 CRAN (R 3.5.0)
scales 0.5.0 2017-08-24 cran (@0.5.0)
scatterplot3d 0.3-41 2018-03-14 cran (@0.3-41)
SDMTools 1.1-221 2014-08-05 cran (@1.1-221)
segmented 0.5-3.0 2017-11-30 cran (@0.5-3.0)
Seurat * 2.3.1 2018-05-05 url
sfsmisc 1.1-2 2018-03-05 cran (@1.1-2)
slam 0.1-43 2018-04-23 CRAN (R 3.5.0)
snow 0.4-2 2016-10-14 cran (@0.4-2)
sparsesvd 0.1-4 2018-02-15 CRAN (R 3.5.0)
splines * 3.5.0 2018-06-18 local
stats * 3.5.0 2018-06-18 local
stats4 * 3.5.0 2018-06-18 local
stringi 1.2.4 2018-07-20 cran (@1.2.4)
stringr * 1.3.1 2018-05-10 CRAN (R 3.5.0)
survival 2.42-6 2018-07-13 CRAN (R 3.5.0)
tclust 1.4-1 2018-05-24 cran (@1.4-1)
tibble * 1.4.2 2018-01-22 cran (@1.4.2)
tidyr * 0.8.1 2018-05-18 cran (@0.8.1)
tidyselect 0.2.4 2018-02-26 cran (@0.2.4)
tidyverse * 1.2.1 2017-11-14 CRAN (R 3.5.0)
timeDate 3043.102 2018-02-21 cran (@3043.10)
tools 3.5.0 2018-06-18 local
trimcluster 0.1-2.1 2018-07-20 cran (@0.1-2.1)
tsne 0.1-3 2016-07-15 cran (@0.1-3)
tweenr 0.1.5 2016-10-10 CRAN (R 3.5.0)
units 0.6-0 2018-06-09 CRAN (R 3.5.0)
utils * 3.5.0 2018-06-18 local
VGAM * 1.0-5 2018-02-07 cran (@1.0-5)
viridis 0.5.1 2018-03-29 cran (@0.5.1)
viridisLite 0.3.0 2018-02-01 cran (@0.3.0)
whisker 0.3-2 2013-04-28 CRAN (R 3.5.0)
withr 2.1.2 2018-03-15 CRAN (R 3.5.0)
workflowr 1.1.1 2018-07-06 CRAN (R 3.5.0)
xml2 1.2.0 2018-01-24 CRAN (R 3.5.0)
yaml 2.2.0 2018-07-25 cran (@2.2.0)
zoo 1.8-3 2018-07-16 cran (@1.8-3) This reproducible R Markdown analysis was created with workflowr 1.1.1